Abstract
Mutations in the zinc finger transcription factor ZIC3 cause X-linked heterotaxy and have also been identified in patients with isolated congenital heart disease (CHD). To determine the relative contribution of ZIC3 mutations to both heterotaxy and isolated CHD, we screened the coding region of ZIC3 in 194 unrelated patients, including 61 patients with classic heterotaxy, 93 patients with heart defects characteristic of heterotaxy, and 11 patients with situs inversus totalis. Five novel ZIC3 mutations in three classic heterotaxy kindreds and two sporadic CHD cases were identified. None of these alleles was found in 97 ethnically matched control samples. On the basis of these analyses, we conclude that the phenotypic spectrum of ZIC3 mutations should be expanded to include affected females and CHD not typical for heterotaxy. This screening of a cohort of patients with sporadic heterotaxy indicates that ZIC3 mutations account for ∼1% of affected individuals. Missense and nonsense mutations were found in the highly conserved zinc finger–binding domain and in the N-terminal protein domain. Functional analysis of all currently known ZIC3 point mutations indicates that mutations in the putative zinc finger DNA binding domain and in the N-terminal domain result in loss of reporter gene transactivation. It is surprising that transfection studies demonstrate aberrant cytoplasmic localization resulting from mutations between amino acids 253–323 of the ZIC3 protein, indicating that the pathogenesis of a subset of ZIC3 mutations results at least in part from failure of appropriate nuclear localization. These results further expand the phenotypic and genotypic spectrum of ZIC3 mutations and provide initial mechanistic insight into their functional consequences.
Introduction
The failure to correctly establish left-right patterning during embryogenesis results in the clinical phenotype of heterotaxy. Affected patients typically have an abnormal arrangement of thoracic and visceral organs, and significant morbidity and mortality result from a wide variety of congenital defects. Complex congenital cardiovascular malformations (CCVM) are classically associated with heterotaxy and account for much of the clinical severity. The incidence of heterotaxy has been difficult to estimate, in part because birth-defect registries track the constellation of anatomic abnormalities separately. In addition, there is no widespread agreement regarding the minimal diagnostic criteria necessary to fulfill a diagnosis of heterotaxy. A recent study with strict diagnostic criteria estimates the prevalence as 1/10,000 births (Lin et al. 2000), closely approximating the findings of the Baltimore-Washington Infant Study, in which the incidence of cardiac malformations associated with abnormal laterality is estimated at 1.44/10,000 live births (Ferencz 1997). In both studies, a male-to-female ratio of ∼2:1 was observed. Overall, this class of defects accounts for ∼3% of congenital heart disease (CHD).
Classic heterotaxy, or situs ambiguus, may include failure of internal organs to lateralize, failure of paired organ primordia to regress, mirror-image reversals, and/or isomerism (Casey 1999; Aylsworth 2001). Two broad clinical categories of heterotaxy—polysplenia and asplenia—are typically used to classify the disorder, although the spleen is not infrequently normal in position and number in heterotaxy cases. Asplenia represents a failure of left-sided patterning and suggests bilateral right-sidedness. Polysplenia suggests bilateral left-sidedness, resulting from either a failure of the midline barrier or a lack of right-side patterning (Peoples et al. 1983; Phoon and Neill 1994; Ticho et al. 2000). Cardiovascular malformations in heterotaxy with asplenia include malposition and transposition of the great arteries (80% of patients), common atrium or atrial septal defects (90% of patients), atrioventricular septal defect (80% of patients), bilateral superior vena cava (50% of patients), pulmonary atresia and stenosis (80% of patients), single ventricle (50% of patients), and total anomalous pulmonary venous connection (70% of patients) (Gutgesell 1998). Several cardiovascular malformations are particularly associated with the polysplenia-type anatomy but are rare in asplenia, including partial anomalous pulmonary venous return (40% of patients), intrahepatic interruption of the inferior vena cava with connection to the azygous or hemiazygous vein (70% of patients), and left ventricular outflow tract obstruction (40% of patients). In addition to cardiac defects, some or all of the abdominal organs may be aberrantly positioned. Common defects include left-sided liver, right-sided stomach, and gut malrotation. Lung lobation may be isomeric or reversed, indicating a failure of correct left-right specification. Although the anatomic constellation of findings consistent with classic heterotaxy has been well documented, laterality disorders likely comprise a spectrum of severity such that milder cases may fail to be ascertained or to be properly identified as left-right patterning defects.
Despite the difficulties associated with clinical and epidemiological studies of heterotaxy, the genetic basis of this disorder has been identified in a small number of familial cases, and studies of left-right patterning in animal models are yielding insights into the underlying molecular pathogenesis (Mercola 1999; Burdine and Schier 2000; Mercola and Levin 2001; Hamada et al. 2002; Bisgrove et al. 2003; Ware and Belmont 2003). Mutations in three genes that function in the transforming growth factor–β (TGFβ) signaling pathway—activin receptor type IIB (ACVR2B [MIM 602730]), an EGF-CFC family member CRYPTIC (MIM 605194), and LEFTYA (MIM 601877)—have been found in a small number of patients with classic heterotaxy (Kosaki et al. 1999a, 1999b; Bamford et al. 2000). Recently, mutations in CRELD1 (MIM 607170), a cell adhesion molecule, and NKX2.5 (MIM 004387) have been associated with heterotaxy (Watanabe et al. 2002; Robinson et al. 2003). However, only single patients with laterality defects were observed in the latter studies, and the phenotypic consequences of gene defects in these loci therefore require further characterization.
The first mutations identified in patients with heterotaxy were in the zinc finger transcription factor ZIC3 (MIM 306955). To date, point mutations have been described in four X-linked familial cases, in one sporadic case of heterotaxy, and in one case of isolated CHD (i.e., congenital heart defect consistent with heterotaxy without other visceral anomalies) (Casey et al. 1993; Ferrero et al. 1997; Gebbia et al. 1997; Megarbane et al. 2000). These alleles include frameshift, missense, and nonsense mutations. In addition, deletion of the ZIC3 locus has been associated with situs ambiguus, suggesting that loss of function may underlie the pathogenesis in the patients with point mutations (Ferrero et al. 1997). Female carriers in the reported familial cases had no CHD, although females in one family had situs inversus with anal stenosis. In addition, ZIC3 truncation has been reported in one family with apparently isolated CHD (Megarbane et al. 2000), suggesting that an unknown proportion of isolated CHD may be caused by mutations in ZIC3. Similarly, mutations in CRYPTIC have recently been identified in patients with isolated transposition of the great arteries (TGA) and double outlet right ventricle (Goldmuntz et al. 2002). These findings have suggested that isolated CHD may be caused by genes involved in left-right axis development and that there may be variable expression of mutant alleles within and between families. It is currently unclear to what degree ZIC3 mutations contribute to sporadic heterotaxy, and this information is necessary to provide accurate counseling information to families.
In the present study, we have undertaken screening for ZIC3 mutations to determine the relative contribution of this gene to sporadic heterotaxy and isolated CHD. Newly identified familial cases have also been analyzed. Novel ZIC3 mutations have been identified and, along with previously reported mutations, their functional significance has been confirmed by transactivation and subcellular-localization assays. These results permit an expansion of the known genotypic and phenotypic spectrum of ZIC3 mutations, provide counseling information for individuals with classic heterotaxy (as well as those with isolated congenital heart defects), and provide insight into the pathophysiology of ZIC3 defects.
Patients and Methods
Patients
For the purposes of the present study we categorized patients with laterality disorders into three subgroups: classic heterotaxy (familial and sporadic), CHD heterotaxy (familial and sporadic), and situs inversus (sporadic). Patients with classic heterotaxy have CCVM and evidence of disrupted left-right patterning in at least one other organ system, including any of the following anomalies: asplenia, polysplenia, evidence of splenic dysfunction, malrotation, omphalocele, abdominal situs inversus, bilateral bilobed lungs, bilateral trilobed lungs, abnormalities of the liver or gallbladder, or stomach situs. Patients were categorized as having CHD heterotaxy if they had normal situs in other organs or if insufficient data were available to make a determination. The types of CCVM included in this study were based on those described in the Baltimore-Washington Infant Study, which included laterality and looping defects. Patients categorized as affected by situs inversus had situs inversus totalis. The disorder was considered familial if a first-degree relative had a congenital heart defect of any type or if a second- or third-degree relative had evidence of a laterality disorder. Further details on enrollment criteria can be obtained at the Baylor College of Medicine Cardiovascular Genetics Web site. Controls were from the Baylor Polymorphism Resource, a set of lymphoblastoid cell lines developed from healthy adults in which ethnic information has been retained.
Phenotypic information was established via patient history and chart review, when available. Informed consent was obtained from all individuals in accordance with the protocol approved by the institutional review board of Baylor College of Medicine. Blood samples were collected from patients and parents, and Epstein-Barr virus–transformed cell lines were established. Genomic DNA was prepared from cell lines or fibroblast cultures (fetal samples) by standard procedures.
Mutation Detection
Exons 1–3 of the ZIC3 gene (GenBank accession number NM_008413) were amplified by PCR, using the following five primer sets: exon 1aF 5′ TCTTGCAGTGACGGAAAGTT-3′ and exon 1aR 5′-TTCATGTTCATGGGGCTGTA-3′; exon 1bF 5′-GCGCACGATCTATCTTCAGG-3′ and exon 1bR 5′-CCGCATATAACGGAAGAAGG-3′; exon 1cF 5′-TACAGCCCCATGAACATGAA-3′ and exon 1cR 5′-TCTCCCTCGCGCTCTAAGT-3′; exon 2F 5′-GCTGCTTGCCTCTGAGAAAC-3′ and exon 2R 5′-ACGTGGAAGACAGTGGGTTG-3′; and exon 3F 5′-GCTCTTGTTTTTGCTTGCAC-3′ and exon 3R 5′-CATTTCCATCTGATTGGTCTCA-3′. All PCRs were performed using 200 ng genomic DNA, 200 μM dNTPs, and 20 pmol of each primer. Samples were purified using Qiagen PCR purification and were sequenced using an ABI 3700.
Plasmids and Mutagenesis
A 1.4-kb Kpn1-DraI fragment of the human ZIC3 cDNA, encompassing the entire ORF, was subcloned in frame into the KpnI-NotI sites of a pHM6 expression vector (Roche) and into the eGFP vector (Clontech), to create an N-terminal hemagglutinin (HA)–tagged fusion and an N-terminal GFP fusion, respectively. ZIC3 mutations were introduced into the Kpn1-Dra1 fragment via Quik-Change site-directed mutagenesis (Stratagene), were sequenced in their entirety, and were subcloned into pHM6.
Transcriptional Assay
HeLa cells were maintained in α-modified Eagle medium with 2.5% fetal bovine serum and 7.5% newborn calf serum. Cells were transfected using lipofectamine (Roche) according to the manufacturer’s instructions. Cotransfections were performed using HA-ZIC3 constructs and an SV40 luciferase reporter plasmid (pGL3 promoter vector; Promega); results were normalized to a promoterless luciferase vector (pGL3 basic; Promega). A pRL-TK luciferase vector served as a control for transfection efficiency. Cells were harvested 48 h after transfection, and luciferase activities were determined using the Dual Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized to Renilla luciferase activity. All results represent a minimum of four experiments.
Immunofluorescence and Subcellular Localization
HeLa cells were seeded onto glass coverslips at 5 × 105 cells/ml 12–18 h prior to transfection. The HA-ZIC3 expression constructs containing wild-type and mutant ZIC3 were transfected into HeLa cells by use of Lipofectamine (Roche), according to the manufacturer’s instructions. After 24 h, the cells were fixed in 4% paraformaldehyde/PBS on ice for 30 min, washed three times in PBS, and permeabilized in 0.3% Triton X/PBS for 2 min at room temperature. After a brief wash in 0.1% Triton X/PBS, the cells were blocked in 1% BSA/0.1% Triton X in PBS at room temperature for 1 h. The cells were incubated with a 1:250 dilution of an anti-HA rabbit polyclonal antibody (Novus Biologicals) for 2 h at room temperature, washed five times in PBS, incubated 1 h with a 1:500 dilution of Alexa Fluor 594 goat anti-rabbit IgG (Molecular Probes), washed three times in PBS then briefly in distilled water and mounted with DAPI. Cells were analyzed using fluorescence microscopy. At least 100 cells were counted for each construct. Cells were scored as “nuclear,” “nuclear and cytoplasmic,” or “cytoplasmic.”
Results and Discussion
Cohort Analysis
ZIC3 mutation analysis was performed by sequencing in a cohort of 20 familial and 145 sporadic cases of heterotaxy. Twenty-nine patients with atrial septal or ventricular septal defects without evidence of a laterality disorder were used as CHD controls. The 165 patients with heterotaxy included 59 females and 106 males, whereas the control CHD group consisted of 13 males and 16 females. The ethnicity of the patients with heterotaxy included 2% Asian (n=3), 3% African American (n=5), 42% white (n=69), 32% Hispanic (n=53), and 21% other or not determined (n=35).
Characteristics of the cohort are summarized in table 1. Patients with classic heterotaxy were subdivided on the basis of spleen position and number. Asplenia and polysplenia are thought to represent bilateral right-sidedness and bilateral left-sidedness, respectively; although they have overlapping spectra of anatomic defects, each has also been associated with characteristic cardiovascular malformations on the basis of autopsy series (Peoples et al. 1983; Van Praagh et al. 1990; Phoon and Neill 1994; Uemura et al. 1995). In our cohort, the asplenia phenotype is more commonly associated with total anomalous pulmonary venous return and dextrocardia, whereas the polysplenia phenotype is more commonly associated with azygous continuation of the inferior vena cava and levocardia. These findings are consistent with previously published reports and indicate that this subpopulation of our cohort reflects the commonly identified anatomic abnormalities seen in heterotaxy. In general, the types of CCVM in the CHD heterotaxy group are similar, both in complexity and anatomy, to those found in the classic heterotaxy group. There is, however, an increased representation of L-TGA and a decreased representation of anomalous pulmonary venous return in the CHD heterotaxy group.
Table 1.
Cardiac Manifestations of the Heterotaxy Cohort
|
Classic Heterotaxy |
|||||
| CardiacFindinga | Asplenia(N = 28) | Polysplenia(N = 9) | Normal Spleen(N = 5) | Not Determined(N = 19) | CHD heterotaxy(N = 93) |
| Levocardia | 7 (25%) | 8 (89%) | 1 | 10 (53%) | 56 (69%) |
| Mesocardia | 0 | 0 | 0 | 2 (11%) | 1 |
| Dextrocardia | 13 (46%) | 1 (11%) | 4 | 6 (32%) | 24 (30%) |
| HLHS | 3 (11%) | 0 | 0 | 2 (11%) | 5 (6%) |
| PA | 8 (29%) | 1 (11%) | 3 | 7 (37%) | 14 (17%) |
| PS | 13 (46%) | 3 (33%) | 1 | 3 (16%) | 28 (35%) |
| CAVC | 19 (68%) | 4 (44%) | 1 | 1 | 4 (5%) |
| DILV | 1 | 0 | 0 | 0 | 3 (4%) |
| DORV | 8 (29%) | 2 (22%) | 2 | 4 (21%) | 17 (21%) |
| Arch abn | 2 | 0 | 2 | 4 (21%) | 6 (7%) |
| D-TGA | 14 (50%) | 3 (33%) | 2 | 8 (42%) | 28 (35%) |
| L-TGA | 0 | 0 | 2 | 1 | 20 (25%) |
| TAPVR | 10 (36%) | 1 (11%) | 1 | 2 (11%) | 3 (4%) |
| PAPVR | 5 (18%) | 0 | 2 | 1 | 3 (4%) |
| SVC abn | 14 (50%) | 5 (56%) | 2 | 4 (21%) | 5 (6%) |
| IVC abn | 9 (32%) | 7 (78%) | 2 | 2 (11%) | 7 (9%) |
Abn = abnormality; CAVC = complete atrioventricular canal; DILV = double inlet left ventricle; DORV = double outlet right ventricle; D-TGA = D-transposition of the great arteries; HLHS = hypoplastic left heart syndrome; IVC = inferior vena cava; L-TGA = L-transposition of the great arteries; PA = pulmonic atresia; PAPVR = partial anomalous pulmonary venous return; PS = pulmonic stenosis; SVC = superior vena cava; TAPVR = total anomalous pulmonary venous return.
Identification of ZIC3 mutations
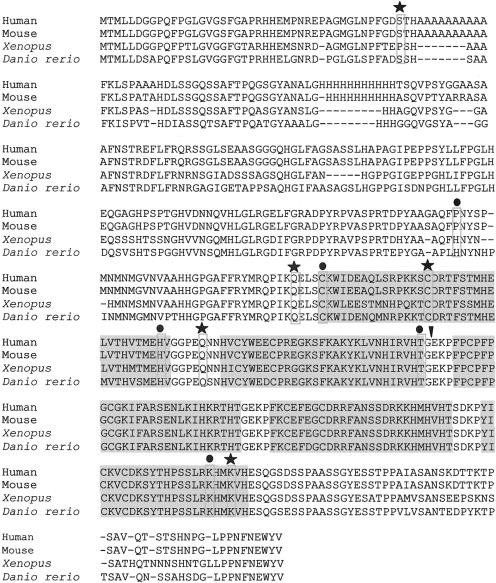
From a total of 194 patient samples, we identified 8 novel ZIC3 nucleotide changes: 2 nonsense mutations, 3 missense mutations, 2 silent nucleotide changes, and 1 nucleotide change in the 3′ untranslated region (table 2). The location of these novel point mutations, as well as those previously reported (Gebbia et al. 1997; Megarbane et al. 2000), and their predicted effects in the ZIC3 amino acid sequence are shown in figure 1.
Table 2.
ZIC3 Nucleotide Changes
| Pedigree | Inheritance | Sex | Ethnicity | NucleotideChange | Amino AcidChange | Amplicon |
| LAT 107 | Familial | M | White | 1262T→A | C253S | 1c |
| LAT 129 | Familial | M | White | 1250C→T | Q249X | 1c |
| LAT 138 | Familial | M | White | 633C→A | S43X | 1a |
| JT 715 | Sporadic | M | White | 1154C→G | P217A | 1b |
| LAT 818 | Sporadic | F | White | 1718A→G | K405E | 2 |
| JT 58 | Sporadic | M | Hispanic | 1417G→A; 1936C→A | R304; R 3′ UTR | 1c; 3 |
| LAT 467 | Sporadic | F | Hispanic | 1366G→A | V287V | 1c |
Figure 1.
Conservation of ZIC3 sequence and location of all known mutations. The five C2H2 zinc finger domains are shaded. The amino acid position for each mutation is boxed. Symbols above each box signify the type of mutation: circle (●) = missense; star (★) = nonsense; downward arrowhead (▾) = insertion.
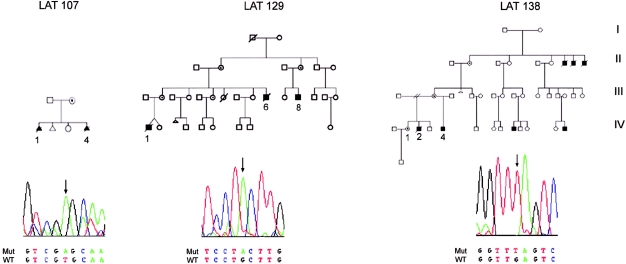
We ascertained three pedigrees with transmission of heterotaxy consistent with X-linked inheritance. ZIC3 was sequenced in the probands (fig. 2), and two nonsense mutations and one missense mutation were identified. In each case, the mother of the proband was heterozygous for the mutation. In pedigree LAT 138, a C→A change results in a nonsense mutation and predicts protein truncation at amino acid 43. All previous mutations have clustered in the zinc finger–binding domains (Gebbia et al. 1997; Megarbane et al. 2000); therefore, this represents the most N-terminal mutation described to date. A second nonsense mutation, located in a conserved glutamine at amino acid 249, was found in pedigree LAT 129. This mutation results in truncation prior to the first zinc finger–binding domain.
Figure 2.
ZIC3 mutations in familial heterotaxy. Sequence traces for the hemizygous proband are shown below each pedigree. Sequences shown for LAT 129 and LAT 138 represent the reverse complement. The phenotypes of affected individuals for whom data are available are shown in table 3.
The third familial mutation, C253S, is located at the first residue of the first zinc finger. Several factors suggest that this missense mutation is pathogenic. First, it is found in the highly conserved cysteine that initiates the first zinc finger domain and that forms one of the C2H2 zinc finger motifs. Second, this change was not found in 145 ethnically matched control chromosomes. Third, it functions abnormally in transactivation and subcellular-localization assays (see below). Figure 1 shows all currently identified ZIC3 mutations, including the five novel mutations identified in the present study, and illustrates the conservation of sequence across species found at the missense mutation sites.
One ZIC3 mutation was identified in the cohort of 145 patients with sporadic heterotaxy (table 2). The mutation, in exon 2, results in the substitution of glutamate for lysine at amino acid 405. This residue is highly conserved (fig. 1) and lies adjacent to a residue that participates in the C2H2 motif at amino acid 406. This change was not found in 145 ethnically matched control chromosomes. It is notable that this is the first female with CHD heterotaxy in whom a ZIC3 mutation has been identified.
The second mutation identified in a patient with nonheterotaxy CHD was from the group of 29 individuals with nonheterotaxy CHD. The patient has an atrial septal defect and pulmonic stenosis (table 3). No other malformation typically associated with heterotaxy was identified. The missense mutation at amino acid 217 results in a substitution of alanine for proline. It is located outside the zinc finger domain in a residue that is incompletely conserved among species (fig. 1). The results of mutation analysis in sporadic cases therefore indicate that ZIC3 mutations account for ∼1% (2/174; 95% CI 0.14%–4.0%) of this patient population. Furthermore, our data indicate that ZIC3 mutations do not underlie the male predominance seen in patients with heterotaxy.
Table 3.
Phenotypic Characteristics of Patients with ZIC3 Mutation [Note]
|
Abnormality in |
||||||||||
| Pedigree | Atria | Ventricles | GreatVessels | AbdominalSitus | Spleen | Gut | Liver/Bile Duct | GU | Skeleton | Other |
| 107 IV-4 | R atrial isomerism | HLHS | TGA; TAPVR | Asplenia | Malrotation | Abnormal liver lobation | Webbed neck; bilat trilobed lungs | |||
| 107 IV-1 | Omphalocele | Horseshoe kidney | Lumbosacral spine anomalies | Cystic hygroma | ||||||
| 129 IV-1 | Situs solitus | Single ventricle; CAVC; PS; DORV | PDA; D-TGA; TAPVR | Abnormal | Asplenia | Horseshoe kidney | Low-set ears | |||
| 129 III-6 | Common atrium | HLHS; PA; dextro; CAVC | Bilat SVC | Asplenia | Tracheoesophageal fistula | Imperforate anus; renal dysplasia | Radial dysplasia; vertebral defect | |||
| 129 III-8 | PS; DORV; VSD | Abnormal IVC; right aortic arch | Bowel obstruction | Cholelithiasis; recurrent cholecystitis | ||||||
| 138 IV-4 | CAVC; DORV | TGA; TAPVR; bilat SVC | Abnormal | Asplenia | Cholestasis with biliary atresia | |||||
| 138 IV-2 | ASD; situs solitus | DORV; VSD; PS | TGA; interrupted IVC | Normal | Normal | Normal | EHBA; cholestatic liver disease | Imperforate anus | Bilat club feet; fused lumbar vertebrae | Posterior embryotoxon; low-set ears |
| 138 IV-1 | Club foot | |||||||||
| 818 | ASD | VI; VSD; PS | L-TGA | |||||||
| 15 | ASD | PS | ||||||||
Note.— Bilat = bilateral; EHBA = extrahepatic biliary atresia; PDA = patent ductus arteriosus. For an explanation of other abbreviations used, please see table 1, footnote a.
Expansion of the Phenotypic Spectrum Associated with ZIC3 Mutations
Table 3 summarizes the phenotypes of the three familial cases (LAT 107, LAT 129, and LAT 138) and the two sporadic cases (LAT 818 and JT 15) in which ZIC3 mutations were identified. For each pedigree, the proband is listed first, followed by other affected family members for whom data were available. Although the familial cases demonstrate anatomic anomalies very consistent with findings in heterotaxy, they exhibit a number of abnormalities not previously described in association with ZIC3 mutations. Novel findings regarding cardiac lesions include two cases of hypoplastic left heart and two cases of total anomalous pulmonary venous return. Novel extracardiac anomalies include omphalocele. Midline defects are commonly associated with disorders of laterality, most frequently affecting genitourinary or musculoskeletal development (Gebbia et al. 1997; Ticho et al. 2000). Genitourinary abnormalities, particularly imperforate anus, have been consistently reported in families with ZIC3 mutations (Casey et al. 1993; Gebbia et al. 1997). In concordance with this, two of the families identified in this study had affected members with imperforate anus. A variety of skeletal and renal defects were noted as well. In addition, a number of features not typically characteristic of heterotaxy syndromes, including dysmorphic facial features, cystic hygroma, and posterior embryotoxon were noted. It is interesting to note that patient LAT 138 IV-2 carried a tentative diagnosis of Alagille syndrome and patient LAT 129 III-6 was given a diagnosis of VATER association (MIM 192350). Because the clinical phenotype of patients with ZIC3 mutations is broad, it should be considered in the differential diagnosis of patients with multiple congenital anomalies, particularly when CCVMs are involved.
It is surprising that the ZIC3 mutations that were found in the subgroup of patients with sporadic CHD were not in males with classic heterotaxy. Both individuals had isolated heart defects without other manifestations of heterotaxy, reinforcing a previous observation that isolated congenital heart defects can result from ZIC3 deficiency (Megarbane et al. 2000). Furthermore, one of the patients was female, and the other patient had a constellation of heart malformations not typically associated with CHD heterotaxy. The finding of an affected female may reflect skewed X inactivation within the proband. However, Zic3-deficient mice, which closely mimic the anatomic abnormalities seen in humans, show left-right patterning abnormalities in a significant percentage (20%–40%) of heterozygous females (Purandare et al. 2002). Overall, these findings confirm previous speculation that ZIC3 mutations may be found both in females and in males and may be associated with isolated, nonheterotaxy CHD. They also reinforce the need for an understanding of the molecular determinants of looping cardiogenesis. Better understanding, in turn, may lead to a categorization of congenital heart defects more closely related to etiology and may aid the evaluation of potential candidate genes.
Mutations in ZIC3 Alter Reporter Gene Transactivation
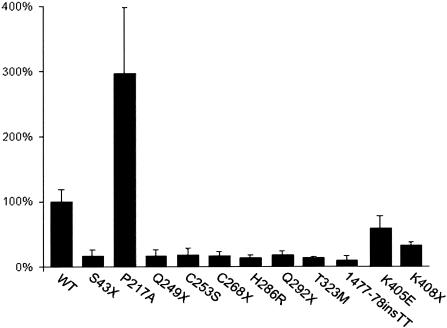
To test the functional significance of mutations in ZIC3, site-directed mutagenesis was used to create previously published and newly identified mutations in the context of the ZIC3 ORF (Gebbia et al. 1997; Megarbane et al. 2000). ZIC3 has been shown to be a weak transcriptional activator and is hypothesized to act as a transcriptional coactivator (Mizugishi et al. 2001). The ZIC family physically and functionally interacts with GLI proteins via the zinc finger–binding domains, but direct transcriptional targets of ZIC3 have not yet been identified in vivo (Koyabu et al. 2001). In vitro, ZIC3 is able to activate a variety of target promoters, including SV40. In HeLa cells, transfection of wild-type HA-tagged ZIC3 demonstrated relatively strong transactivation of an SV40 luciferase reporter, with levels ∼200-fold higher than those of a promoterless control (data not shown). We subsequently tested mutant constructs containing nonsense, missense, or frameshift mutations described in the present study and in others (Gebbia et al. 1997; Megarbane et al. 2000). The results demonstrate that ZIC3 mutations produce aberrant reporter gene transactivation (fig. 3). Loss of activation occurs both with mutations in the putative DNA-binding domain of the zinc finger region as well as with mutations in the N-terminal domain. These results are consistent with previous work in Xenopus, which demonstrated that expression of the N-terminus of Zic3, in the absence of the zinc finger–binding domain, causes left-right patterning defects (Kitaguchi et al. 2000).
Figure 3.
ZIC3 mutations alter transactivation of an SV40 luciferase reporter gene. The luciferase activity of an SV40-luciferase construct cotransfected with HA-ZIC3 wild-type (WT) or mutant constructs is indicated. The specific HA-ZIC3 construct used for cotransfection is shown below each bar. Luciferase activities were measured relative to a promoterless control plasmid and the mean fold-activation as compared with that of the wild type is expressed as a percentage. Each bar represents a minimum of four separate experiments. Vertical lines indicate the standard deviation for each set of experiments.
The nonsense mutations all show significant loss of activation, including a 1477–1478insTT frameshift mutation that results in a premature stop codon at amino acid 408. All but one of the missense mutations also shows a loss of transactivation. The exception is the N-terminal mutation, P217A. In this case, a significant and reproducible increase in transcriptional activation is noted. This mutation was not found in 145 ethnicity-matched control chromosomes or in 412 chromosomes within the CHD cohort. It is of interest that the phenotype of this patient, which consisted of an atrial septal defect and pulmonic stenosis, is not characteristic of heterotaxy (table 3). It is therefore interesting to speculate that activation of ZIC3 may contribute to additional phenotypic abnormalities separable from classic heterotaxy. Zic1 has previously been shown to play a role in the subcellular localization, and thus posttranslational control, of Gli1 and Gli3 in a cell-type–specific manner. Given the known physical interaction between ZIC and GLI proteins, it is possible that increases or decreases in ZIC3 expression alter the stoichiometry of the GLI proteins at the cellular level. Further mutation and functional analyses, both in vitro and in vivo, will be required to establish whether activating mutations or overexpression of the ZIC3 transcription factor plays a significant role in CHD.
Subcellular Localization
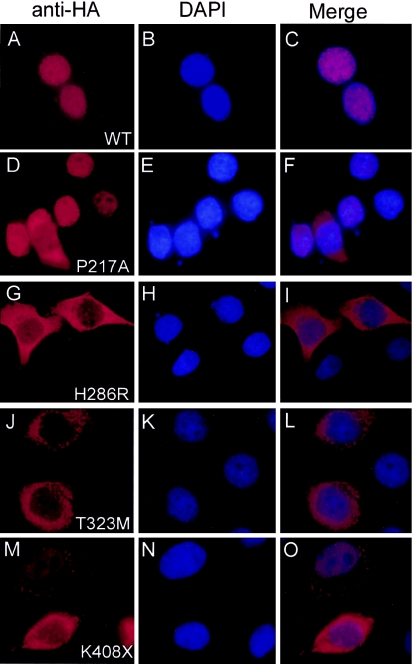
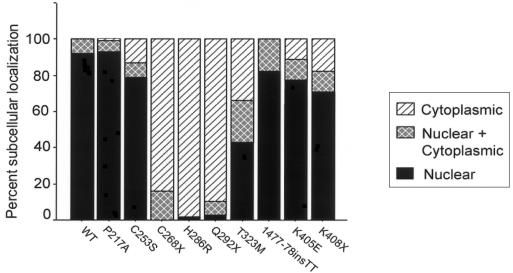
To further assess the functional significance of ZIC3 mutations, transient transfections were performed using ZIC3-mutant constructs to determine subcellular localization by use of immunofluorescence. Initially, the appropriate expression pattern of ZIC3 and the effect of epitope tagging were assessed using two constructs, an N-terminal HA tagged ZIC3 and an N-terminal eGFP-ZIC3 fusion. These constructs demonstrated nuclear localization of ZIC3 in both HeLa and P19 teratocarcinoma cells (fig. 4 and data not shown), and HA-tagged constructs were used for the remainder of the experiments. It is interesting that all of the missense mutations tested, with the exception of P217A, showed abnormal subcellular localization (fig. 4 and data not shown). HA-ZIC3 was detected in the cytoplasm or in both cytoplasm and nucleus. In some cells, localization within the cytoplasm was in a stippled distribution. To compare the relative distribution of ZIC3 by construct, 100–200 cells were counted for each construct and were scored for nuclear, nuclear and cytoplasmic, or cytoplasmic localization. The results are shown in figure 5 and indicate that loss of nuclear localization occurs for mutations between amino acids 253 and 323. Nuclear-localization–prediction programs fail to identify a nuclear localization sequence (NLS) for ZIC3. The lack of a classical NLS suggests that ZIC3 may enter the nucleus as a complex associated with other factors. Alternatively, ZIC3 may contain a novel NLS or may enter the nucleus directly via an unconventional pathway. The results obtained using these known patient mutations delimit a critical protein domain(s) required for nuclear localization. Furthermore, these results indicate that the pathogenesis of a subset of ZIC3 mutations results from failure of the mutant protein to localize to the nucleus.
Figure 4.
Immunofluorescent subcellular localization of ZIC3. For each construct, anti-HA (panels A, D, G, J, and M) and DAPI (panels B, E, H, K, and N) staining are shown individually and merged (panels C, F, I, L, and O). The wild-type construct is located in the nucleus (panels A–C); the missense mutation construct P217A is primarily nuclear (panels D–F), whereas H286R is located in the cytoplasm (panels G–I). T323M (panels J–L) and K408X (panels M–O) show evidence of cytoplasmic stippling.
Figure 5.
Mutations in ZIC3 alter subcellular localization. The subcellular localization of anti-HA staining is shown graphically for each construct. Nuclear, nuclear and cytoplasmic, and cytoplasmic staining are indicated as percentages.
In addition to abnormalities in subcellular localization, mutations also affected protein stability. Two truncating mutations, S43X and Q249X, resulted in absent or nearly absent protein (data not shown). Furthermore, on the basis of the number of HA-expressing cells in this assay, protein stability also appeared to be qualitatively diminished with several of the missense mutations. A summary of the mutations with their associated clinical information and results of functional analyses is shown in table 4.
Table 4.
Summary of Mutation Analyses[Note]
| Mutation | Inheritance, Sex, and Clinical Phenotype | Reporter GeneTransactivation | SubcellularLocalization | MutationReference |
| S43X | Fam; M; H | D | No protein detected | Present study |
| P217A | Spo; M; ASD, PS | I | Nuclear | Present study |
| Q249X | Fam; M; H | D | No protein detected | Present study |
| C253S | Fam; M; H | D | 80% nuclear | Present study |
| C268X | Fam; M; H | D | Cytoplasmic | Gebbia et al. 1997 |
| H286R | Fam; M; H | D | Cytoplasmic | Gebbia et al. 1997 |
| Q292X | Fam; M; H | D | Cytoplasmic | Gebbia et al. 1997 |
| T323M | Fam; M; H | D | 45% nuclear | Gebbia et al. 1997 |
| 1477–1478 ins TT | Fam; M; H | D | >80% nuclear | Gebbia et al. 1997 |
| K405E | Spo; F; isolated CHD | D | 80% nuclear | Present study |
| K408X | Fam; M; isolated CHD | D | 70% nuclear | Megarbane et al. 2000 |
ASD = atrial septal defect; D = decreased; F = female; Fam = familial; H = classic heterotaxy; I = increased; isolated CHD = congenital heart defect consistent with heterotaxy without other visceral anomalies; M = male; PS = pulmonic stenosis; Spo = sporadic.
In summary, our data demonstrate that ZIC3 mutations account for ∼1% of sporadic CHD heterotaxy. The additional mutations identified in the present study indicate that mutations outside the zinc finger–binding domain can cause heterotaxy and further broaden the genotypic and phenotypic spectrum of defects caused by mutations in this gene. In addition, our data provide methods for functional evaluation of ZIC3 mutations and indicate that the pathogenesis of a subset of mutations results from aberrant subcellular localization. Future studies directed toward identifying the interactions required for proper cellular localization should provide valuable insight into the mechanisms by which ZIC3 functions in left-right patterning.
Acknowledgments
We thank all the families and patients for their participation. We thank Laura Molinari in the Tissue Culture Core; Ana Combes and Andres Menesses-Diaz, for patient enrollment; Trang Ho-Dawson, for bioinformatic support,; and Ms. Meg Hefner, Dr. Gautam Singh, Ms. Karlene Coleman, Dr. Jonathan Zonana, and Ms. Karen Kovak, for referring patients with familial heterotaxy described in this study. This work was supported by National Institutes of Health grants HL67355 and HD41648 (to S.M.W.) and grants HD39056 and HL067155 (to J.W.B.).
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Baylor College of Medicine Cardiovascular Genetics, http://www.cardiogene.org (for details on enrollment criteria)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ZIC3 [accession number NM_003413])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ZIC3, CRYPTIC, ACVR2B, LEFTYA, CRELD1, NKX2.5, and VATER)
References
- Aylsworth AS (2001) Clinical aspects of defects in the determination of laterality. Am J Med Genet 101:345–355 10.1002/ajmg.1219.abs [DOI] [PubMed] [Google Scholar]
- Bamford RN, Roessler E, Burdine RD, Saplakoglu U, de la Cruz J, Splitt M, Goodship JA, Towbin J, Bowers P, Ferrero GB, Marino B, Schier AF, Shen MM, Muenke M, Casey B (2000) Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet 26:365–369 10.1038/81695 [DOI] [PubMed] [Google Scholar]
- Bisgrove B, Morelli S, Yost H (2003) Genetics of human laterality disorders: insights from vertebrate model systems. Annu Rev Genomics Hum Genet 4:1–32 10.1146/annurev.genom.4.070802.110428 [DOI] [PubMed] [Google Scholar]
- Burdine RD, Schier AF (2000) Conserved and divergent mechanisms in left-right axis formation. Genes Dev 14:763–776 [PubMed] [Google Scholar]
- Casey B (1999) Genetics of human left-right axis malformations. In: Harvey RP, Rosenthal N (eds) Heart development. San Diego, Academic Press, pp 479–489 [Google Scholar]
- Casey B, Devoto M, Jones K, Ballabio A (1993) Mapping a gene for familial situs abnormalities to human chromosome Xq24–q27.1. Nat Genet 5:403–407 [DOI] [PubMed] [Google Scholar]
- Ferencz C, Loffredo CA, Correa-Villasenor A, Wilson PD (eds) (1997) Defects of laterality and looping. Futura Publishing, Armonk, NY [Google Scholar]
- Ferrero GB, Gebbia M, Pilia G, Witte D, Peier A, Hopkin RJ, Craigen WJ, Shaffer LG, Schlessinger D, Ballabio A, Casey B (1997) A submicroscopic deletion in Xq26 associated with familial situs ambiguus. Am J Hum Genet 61:395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, Nelson DL, Casey B (1997) X-linked situs abnormalities result from mutations in ZIC3. Nat Genet 17:305–308 [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Bamford R, Karkera JD, de la Cruz J, Roessler E, Muenke M (2002) CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet 70:776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutgesell HP (1998) Cardiac malposition and heterotaxy. In: Garson AJ, Bricker JT, Fisher DJ, Neish SR (eds). The science and practice of pediatric cardiology. Vol 2. Williams & Wilkins, Baltimore, pp 1539–1563 [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y (2002) Establishment of vertebrate left-right asymmetry. Nat Rev Genet 3:103–113 10.1038/nrg732 [DOI] [PubMed] [Google Scholar]
- Kitaguchi T, Nagai T, Nakata K, Aruga J, Mikoshiba K (2000) Zic3 is involved in the left-right specification of the Xenopus embryo. Development 127:4787–4795 [DOI] [PubMed] [Google Scholar]
- Kosaki K, Bassi MT, Kosaki R, Lewin M, Belmont J, Schauer G, Casey B (1999a) Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet 64:712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, Casey B (1999b) Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet 82:70–76 [DOI] [PubMed] [Google Scholar]
- Koyabu Y, Nakata K, Mizugishi K, Aruga J, Mikoshiba K (2001) Physical and functional interactions between Zic and Gli proteins. J Biol Chem 276:6889–6892 10.1074/jbc.C000773200 [DOI] [PubMed] [Google Scholar]
- Lin AE, Ticho BS, Houde K, Westgate MN, Holmes LB (2000) Heterotaxy: associated conditions and hospital-based prevalence in newborns. Genet Med 2:157–172 10.1097/00125817-200005000-00002 [DOI] [PubMed] [Google Scholar]
- Megarbane A, Salem N, Stephan E, Ashoush R, Lenoir D, Delague V, Kassab R, Loiselet J, Bouvagnet P (2000) X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur J Hum Genet 8:704–708 10.1038/sj.ejhg.5200526 [DOI] [PubMed] [Google Scholar]
- Mercola M (1999) Embryological basis for cardiac left-right asymmetry. Semin Cell Dev Biol 10:109–116 10.1006/scdb.1998.0280 [DOI] [PubMed] [Google Scholar]
- Mercola M, Levin M (2001) Left-right asymmetry determination in vertebrates. Annu Rev Cell Dev Biol 17:779–805 10.1146/annurev.cellbio.17.1.779 [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Aruga J, Nakata K, Mikoshiba K (2001) Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem 276:2180–2188 10.1074/jbc.M004430200 [DOI] [PubMed] [Google Scholar]
- Peoples WM, Moller JH, Edwards JE (1983) Polysplenia: a review of 146 cases. Pediatr Cardiol 4:129–137 [DOI] [PubMed] [Google Scholar]
- Phoon CK, Neill CA (1994) Asplenia syndrome: insight into embryology through an analysis of cardiac and extracardiac anomalies. Am J Cardiol 73:581–587 [DOI] [PubMed] [Google Scholar]
- Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B (2002) A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development 129:2293–2302 [DOI] [PubMed] [Google Scholar]
- Robinson SW, Morris CD, Goldmuntz E, Reller MD, Jones MA, Steiner RD, Maslen CL (2003) Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet 72:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticho BS, Goldstein AM, Van Praagh R (2000) Extracardiac anomalies in the heterotaxy syndromes with focus on anomalies of midline-associated structures. Am J Cardiol 85:729–734 10.1016/S0002-9149(99)00849-8 [DOI] [PubMed] [Google Scholar]
- Uemura H, Ho SY, Devine WA, Anderson RH (1995) Analysis of visceral heterotaxy according to splenic status, appendage morphology, or both. Am J Cardiol 76:846–849 10.1016/S0002-9149(99)80243-4 [DOI] [PubMed] [Google Scholar]
- Van Praagh S, Kreutzer J, Alday L, Van Praagh R (1990) Systemic and pulmonary venous connections in visceral heterotaxy, with emphasis on the diagnosis of the atria situs: a study of 109 postmortem cases. In: Clark E, Takao A (eds). Developmental cardiology: morphogenesis and function. Futura, Mt. Kisco, NY [Google Scholar]
- Ware S, Belmont J (2003) Tgfβ signaling in midline and laterality defects. In: Wynshaw-Boris A (ed) Inborn errors of development: the molecular basis of clinical disorders of morphogenesis. New York, Oxford University Press [Google Scholar]
- Watanabe Y, Benson DW, Yano S, Akagi T, Yoshino M, Murray JC (2002) Two novel frameshift mutations in NKX2.5 result in novel features including visceral inversus and sinus venosus type ASD. J Med Genet 39:807–811 10.1136/jmg.39.11.807 [DOI] [PMC free article] [PubMed] [Google Scholar]