Abstract
The coronavirus replicase gene (gene 1) is translated into two co-amino-terminal polyproteins that are proteolytically processed to yield more than 15 mature proteins. Several gene 1 proteins have been shown to localize at sites of viral RNA synthesis in the infected cell cytoplasm, notably on late endosomes at early times of infection. However, both immunofluorescence and electron microscopic studies have also detected gene 1 proteins at sites distinct from the putative sites of viral RNA synthesis or virus assembly. In this study, mouse hepatitis virus (MHV)-infected cells were fractionated and analyzed to determine if gene 1 proteins segregated to more than one membrane population. Following differential centrifugation of lysates of MHV-infected DBT cells, gene 1 proteins as well as the structural N and M proteins were detected almost exclusively in a high-speed small membrane pellet. Following fractionation of the small membrane pellet on an iodixanol density gradient, the gene 1 proteins p28 and helicase cofractionated with dense membranes (1.12 to 1.13 g/ml) that also contained peak concentrations of N. In contrast, p65 and p1a-22 were detected in a distinct population of less dense membranes (1.05 to 1.09 g/ml). Viral RNA was detected in membrane fractions containing helicase, p28, and N but not in the fractions containing p65 and p1a-22. LAMP-1, a marker for late endosomes and lysosomes, was detected in both membrane populations. These results demonstrate that multiple gene 1 proteins segregate into two biochemically distinct but tightly associated membrane populations and that only one of these populations appears to be a site for viral RNA synthesis. The results further suggest that p28 is a component of the viral replication complex whereas the gene 1 proteins p1a-22 and p65 may serve roles during infection that are distinct from viral RNA transcription or replication.
The genome of the coronavirus mouse hepatitis virus (MHV) is a 32-kb single-stranded positive-sense RNA molecule. The nonstructural proteins required for virus replication and transcription are encoded within gene 1. Gene 1 comprises the 5′-most two-thirds (22 kb) of the genome and contains two overlapping open reading frames, ORF1a and ORF1b (6, 10, 30, 35) (Fig. 1). Translation of ORF1b requires a ribosomal frameshifting event at the 3′ end of ORF1a (11), and translation of gene 1 results in two co-amino-terminal polyproteins, pp1a (495 kDa) and pp1ab (803 kDa). At least 15 proteins have been identified as cleavage products of the MHV gene 1 polyproteins. Cleavage of the amino-terminal proteins, p28 and p65, is mediated by the papain-like proteinase 1 (PLP-1) (7, 8, 19, 21, 30). The 3C-like proteinase (3CLpro) has been confirmed or predicted to cleave 11 proteins, including the putative RNA-dependent RNA polymerase (Pol) and helicase (Hel) (30, 31, 33, 34), as well as a recently identified cassette of four small proteins encoded by ORF1a carboxy terminal to 3CLpro (9, 32). The best characterized of these proteins is a 22-kDa cleavage product, p1a-22, which, along with the other three proteins, is significantly conserved among the coronaviruses (9). However, with the exception of the experimentally confirmed viral proteinases and the predicted Pol and Hel proteins, the functions of the mature gene 1 proteins have not been determined.
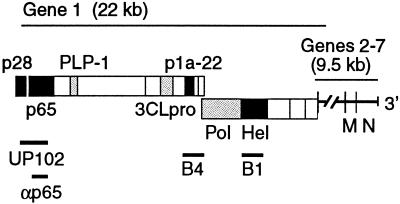
FIG. 1.
MHV genome organization, gene 1 polyprotein processing products, and antibodies. The locations of genes 1 through 7 are shown above the schematic. Protein domains of confirmed or predicted mature gene 1 proteins are shown as boxes. Grey boxes indicate the viral proteinases (PLP-1 and 3CLpro) and the putative RNA-dependent RNA Pol. Black boxes indicate proteins of interest in the current study: the amino-terminal cleavage products, p28 and p65; p1a-22, a small protein carboxy terminal to 3CLpro; and the putative RNA Hel. Lines below the schematic indicate the cloned regions used to generate polyclonal rabbit antisera against gene 1 proteins: UP102 (anti-p28/p65), B4 (anti-p1a-22), B1 (anti-Hel), and anti-p65. Also shown are the locations of genes 6 and 7, encoding the structural membrane (M) and nucleocapsid (N) proteins, respectively.
Gene 1 proteins have been shown to localize to cytoplasmic, predominantly perinuclear foci by indirect immunofluorescence (IF) microscopy (5). Confocal IF microscopy studies using dual labeling of gene 1 and structural proteins have demonstrated that gene 1 proteins colocalize in cytoplasmic membranous complexes along with the nucleocapsid protein (N) (9, 17). Viral RNA synthesis has been detected on intracellular membranes (18) and has been shown to colocalize with gene 1 proteins including the viral Hel and N by confocal microscopy (5, 17, 37). Recent immunoelectron microscopy (immuno-EM) studies of MHV-A59-infected cells have shown that at early times of infection the gene 1-encoded Pol and p1a-22 localize to membranes derived from late endosomes, as does viral RNA (40). Immuno-EM studies also indicated that individual gene 1 proteins may target different intracellular structures, although the relationship of these structures and their roles in RNA synthesis could not be determined based on ultrastructural studies alone. Most recently, it was determined by confocal microscopy that at later times of infection the small ORF1a-encoded protein, p1a-22, was detectable in foci that were distinct from but closely approximate to the complexes containing other gene 1 proteins and viral RNA synthesis (9). Together, these results suggest that gene 1 proteins may target more than one distinct membrane population that may serve independent functions during viral replication (9).
In this report, lysates of metabolically labeled, MHV-infected DBT cells were fractionated and analyzed to determine if biochemically distinct membrane populations containing gene 1 and structural proteins and viral RNA were formed during the course of infection. This study demonstrates that, in a population of MHV-infected cells, gene 1 proteins that appear to colocalize by confocal IF or immuno-EM imaging in fact segregate into at least two distinct membrane populations. Although both gene 1 protein-containing membrane populations also contained markers for endosomes-lysosomes, only the membrane fractions containing p28, Hel, and the structural N contained viral RNA. These studies indicate that gene 1 proteins cleaved from the same polyprotein are able to target closely approximate but biochemically distinct membranes and perhaps serve functions independent from RNA synthesis.
MATERIALS AND METHODS
Virus, cells, and antisera.
DBT cell monolayers were infected with MHV-A59 in all experiments (24). All antisera used for biochemical experiments have been previously published. These antisera include UP102 (anti-p28/anti-p65) (14), B4 (CT1a, anti-p1a-22) (32), and B1 (anti-Hel) (17) for gene 1 proteins. Two monoclonal antisera directed against the structural proteins, nucleocapsid (anti-N, J.3.3) and matrix-membrane (anti-M, J.1.3) were generously provided by John Fleming (University of Wisconsin, Madison). Anti-LAMP-1 antiserum (1D4B) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa) (12).
Anti-p65 antiserum was generated against an amino-terminal domain of p65 overexpressed in Escherichia coli for immunization of New Zealand White rabbits. All immunizations were performed by Cocalico, Inc. Reverse transcription-PCRs were performed using MHV-A59 genome RNA as template. All nucleotide and amino acid numbers correspond to the MHV-A59 sequence modified by Bonilla et al. (6). The p65 PCR product spanned nucleotides 951 to 1776 (amino acids V248 to L521), and a primer-generated restriction site was used to introduce an NcoI site on the 5′ end. The XhoI site at nucleotide 1770 was used as the 3′ site. The p65 fragment was subcloned into the pET23d vector (Novagen) to produce a six-histidine-tagged protein for purification by nickel resin chromatography and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) electroelution.
Infection, radiolabeling, and cell lysis.
DBT cells plated in five 150-cm dishes (5 × 108 cells) were mock infected or infected (multiplicity of infection [MOI] of 10) in Dulbecco modified Eagle medium (DMEM) that contained 10% fetal calf serum (FCS). At 2.5 h postinfection (p.i.), the medium was replaced with DMEM lacking methionine and cysteine with 2% FCS and 10 μg of actinomycin D per ml. The FCS was not dialyzed and thus contained some residual methionine-cysteine. We have previously demonstrated that cells are capable of supporting full translation, replication, and metabolic labeling under these conditions (25) and that the presence of actinomycin D starting at more than 2 h p.i. does not affect cell morphology for up to 10 h. At 6 h p.i., cells were washed with 1× Tris, trypsinized, washed, pelleted, and labeled in suspension for 1 h with DMEM–2% FCS minus methionine-cysteine, actinomycin D, and 100 μCi of [35S]methionine-cysteine (total volume of 2 ml) for 1 h at 37°C. Following the labeling, cells were pelleted and washed once with ice-cold phosphate-buffered saline (PBS), followed by a wash in cold sucrose-Tris buffer (10 mM Tris [pH 7.2], 200 mM sucrose). All subsequent steps were performed at 4°C. Cells (total volume of cell pellet, ∼200 μl) were resuspended in 800 μl of sucrose-Tris buffer to a final volume of 1 ml. Cells were lysed by 40 passes through a ball-bearing homogenizer (0.1666-mm ball bearing), until more than 90% of the cells were lysed (2, 3). For labeling viral RNA, monolayers of DBT cells were infected at an MOI of 10. At 3 h p.i., the inoculum was removed and fresh DMEM with 2% FCS with 10 μg of actinomycin D per ml was added. At 6 h p.i., cells were harvested by trypsin and again labeled in suspension (total volume of 2 ml) for 2 h with 100 μCi of [3H]uridine at 37°C. Cells were pelleted, washed, and resuspended in sucrose-Tris buffer prior to lysis. Cells were lysed as described above.
Differential centrifugation and gradient fractionation.
Cell lysates from either the [35S]methionine-labeled protein or [3H]uridine-labeled RNA samples were subjected to multiple rounds of differential centrifugation. Cellular nuclei were pelleted for 5 min at 1,000 × g at 4°C, mitochondria were pelleted at 2,300 × g for 10 min at 4°C (P2.3), and the remaining intracellular membranes were pelleted at 100,000 rpm (P100) (320,000 × g) for 15 min at 4°C in a Beckman tabletop ultracentrifuge in a Sorvall rotor (TLA-120). The final supernatant (S100 cytosol) was saved as the soluble cellular material. Resuspended pellets were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
To separate the membranes contained within P100, the pellet was subjected to gradient fractionation. Iodixanol (Optiprep-Nycomed Pharma) was diluted into a 50% stock solution in 10 mM Tris, pH 7.2. Iodixanol gradients were formed by sequential addition of 10, 15, 20, 25, and 30% solutions in sucrose-Tris buffer. For each step of the gradient, 900 μl was used for a final volume of 4.5 ml in a 5-ml tube. After pouring, the gradient was incubated at 4°C overnight, resulting in the formation of a continuous gradient. Five hundred microliters of P100, from either mock-infected or infected samples, was loaded onto the gradient followed by centrifugation at 50,000 rpm (300,000 × g) for 3 h at 4°C (Beckman SW50.1 rotor). Gradients were separated into a total of 10 500-μl fractions. For the labeled RNA experiments, 50 100-μl fractions were obtained. All fractions were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Detection of MHV proteins and viral RNA.
Pellets from differential centrifugation and fractions from the gradients were each subjected to immunoprecipitation reactions to determine where the MHV structural and nonstructural proteins were localized. For each reaction, 180 μl of high-salt TTK buffer (175 mM KCl, 10 mM Tris [pH 7.2], 1% Triton X-100), 2 μl of antisera, and 20 μl of sample were added and incubated on ice for 1 h. Protein A-Sepharose beads were then added to each reaction mixture and agitated on a vortex shaker for 1 h at 4°C. Beads were washed twice at 4°C with 100 μl of low-salt TTK buffer (100 mM KCl, 10 mM Tris [pH 7.2], 1% Triton X-100). After the final wash was removed, 15 μl of 2× Laemmli buffer (29) was added to each sample and incubated at room temperature for 5 min with occasional vortexing, followed by boiling the samples for 5 min prior to loading onto SDS–12% polyacrylamide gels. Gels were processed and assayed by fluorography as previously described (13) or by a Fuji phosphoimager.
Differential centrifugation and gradient fractionation samples from the [3H]uridine-labeled RNA samples were all precipitated with trichloroacetic acid (TCA) to isolate viral RNA. Equal volumes of the pellet or fraction were incubated with 1 ml of ice-cold 8% TCA on ice for 30 min. Following the incubation period, the precipitated solution was vacuumed onto a Whatman glass filter, rinsed twice with ice-cold 5% TCA, and allowed to dry for at least 1 h. Reactions were counted in a Beckman LS 6500 scintillation counter.
Assays for the cell marker proteins.
To test for the presence of membranes derived from the Golgi apparatus, iodixanol fractions were assayed for galactosyltransferase activity by incubation with 50 mM Tris (pH 6.8), 0.2% Triton X-100, 40 mM MnCl2, 1 mg of ovalbumin per ml, 2 mM ATP, and 0.25 μCi of [3H]UDP-galactose for 1 h at 37°C. Reaction solutions were then spotted onto Whatman filter paper and fixed in cold 10% TCA at room temperature for 10 min. Filters were quickly rinsed with water three times and then washed in water three times, 5 min per wash, at room temperature. Filters were dried completely and analyzed by scintillation counting. Total counts were adjusted for protein concentration.
Gradient fractions were assayed for the presence of endoplasmic reticulum (ER) membranes by testing fractions for activity of NADPH-cytochrome c reductase, a resident enzyme of the ER, in contrast to the more commonly assayed NADH-cytochrome c reductase activity of the mitochondria. Samples were mixed in 0.1 M potassium phosphate buffer, pH 7.7, with 45 nM NADPH and 5 nM cytochrome c in a total volume of 1 ml. The change in absorbance over time was read at 550 nm at 25°C for 10 min, adjusted for protein concentration of each sample.
LAMP-1 is a marker for both late endosomes and lysosomes (12). Equal volumes of iodixanol gradient fractions were run on SDS–12% polyacrylamide gels, transferred to nitrocellulose membranes (Bio-Rad), and assessed for LAMP-1 by immunoblotting. Primary antibody (1D4B) was used at a 1:500 dilution, and secondary anti-rabbit horseradish peroxidase-conjugated antibodies (Amersham) were used at a 1:1,000 dilution and developed by chemiluminescence per the manufacturer's instructions (NEN Life Sciences). Band densitometry was measured with NIH Image 1.62 (rsb.info.nih.gov/nih-image).
IF assays.
Studies were performed as previously described (17). Briefly, DBT cells on glass coverslips were mock infected or infected with MHV-A59 at an MOI of 10 for 5.5 h. Cells were fixed in −20°C 100% methanol until use. The cells were rehydrated in PBS and blocked with 5% bovine serum albumin prior to incubation with primary antisera against the structural protein nucleocapsid (J.3.3) or membrane (J.1.3). Cells were then incubated with Cy3-conjugated goat anti-mouse antisera. p65 was then detected using polyclonal anti-p65 rabbit sera, followed by Cy2-conjugated secondary goat anti-rabbit antisera. Dual-labeled cells were imaged at 586 and 488 nm for Cy3 and Cy2, respectively, on a Zeiss LSM 410 confocal microscope. Acquisition of images was optimized in the LSM software, and simultaneous dual imaging was performed to ensure accurate registration. Red (Cy3) and green (Cy2) colors were assigned to all gray-scale images using the hue-saturation option, and images were merged in Adobe Photoshop 5.0.
RESULTS
Membrane association of MHV replicase proteins.
Biochemical fractionation of MHV-infected cells has been used to confirm that coronavirus RNA species are associated with cellular membranes, possibly protected within large complexes (18, 36); however, biochemical fractionation studies of gene 1 proteins have not previously been performed. Since our previous gene 1 protein expression studies and protein localization cell imaging studies were performed with MHV-permissive DBT cells, these cells were also used for all biochemical analyses. To determine if gene 1 proteins were membrane associated, crude differential centrifugation pellets and cytosol from MHV-infected cells were assayed for the presence of MHV gene 1 and structural proteins. Radiolabeled, mock-infected, and infected cells were lysed by ball bearing homogenization (2, 3) and subjected to three rounds of differential centrifugation. The membrane pellets were immunoprecipitated with antibodies to several gene 1 and structural proteins and analyzed by SDS-PAGE and fluorography.
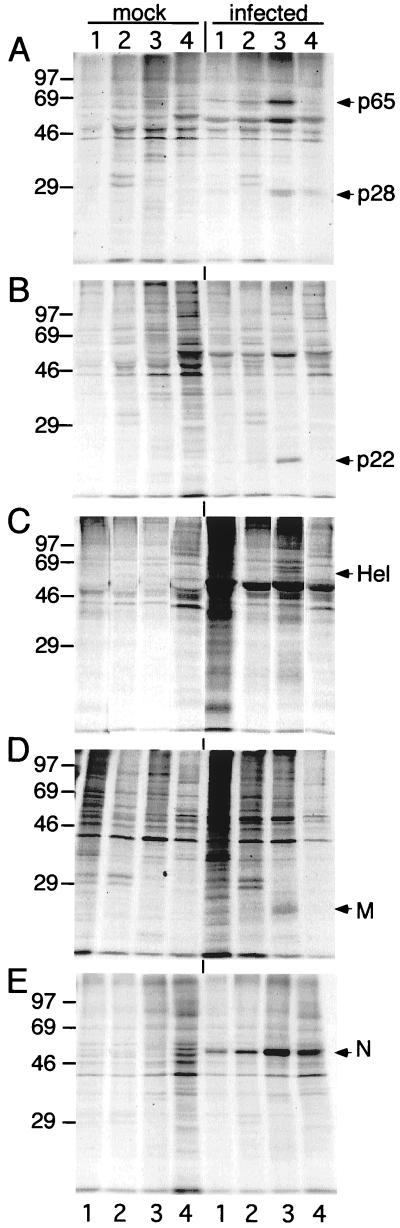
Hel and p1a-22 were detected exclusively in the high-speed membrane pellet (P100) (Fig. 2B and C). p28 and p65 were also predominantly detected in P100; however, a lesser amount of p28 was also detected in the post-P100 supernatant (S100 cytosol) (Fig. 2A). These results demonstrated that all gene 1 proteins tested were associated with intracellular membranes. This experiment also demonstrated that MHV gene 1 proteins could be concentrated in cellular membrane fractions by simple differential centrifugation.
FIG. 2.
Differential centrifugation and immunoprecipitation of lysates from MHV-infected DBT cells. Cells were infected, lysed, spun, and immunoprecipitated as described in Materials and Methods. Proteins were separated on SDS–12% polyacrylamide gels and subjected to fluorography. Marker proteins (molecular masses in kilodaltons) are to the left of each gel, and proteins of interest are indicated to the right of each gel. Gels are aligned so that lane markers and labels on top of panel A also apply to panel B through E. Mock-infected (mock) and infected cell lysates are shown at the top of the gel. Lanes: 1, nuclear pellet (P1; 1,000 × g); 2, P2.3 pellet (2,300 × g); 3, P100 pellet (320,000 × g); 4, supernatant from 320,000 × g spin (S100 cytosol). (A) UP102, anti-p28/p65. (B) B4, anti-p1a-22. (C) B1, anti-Hel. (D) anti-M (J.1.3) monoclonal antibody (E) anti-N (J.3.3) monoclonal antibody.
We next determined the localization of the structural nucleocapsid (N) and membrane (M) proteins. Both M and N were detected in crude membrane pellets, with M detected exclusively in P100, consistent with its known association with membranes of the intermediate compartment and Golgi apparatus (Fig. 2D) (26, 28, 38, 39). N was also predominantly detected in P100, but in contrast to M, N also was detected in the other crude membrane pellets (Fig. 2E) and in the S100 cytosol. Detection of M in only P100 indicated that the detection of N in multiple pellets was not an artifact of homogenization. Instead, it more likely reflected the abundance of N and its ability to bind different intracellular membranes (1). The presence of N in the nuclear pellet was most likely due to the adherence of membranes to the nuclei following gentle homogenization. N was not detected with nuclei when the pelleted nuclei were resuspended in buffer containing NP-40 and then repelleted (data not shown). Thus, all of the tested MHV proteins were either predominantly or exclusively detected in the P100 pellet, indicating principal association with membranes of late endosomes, lysosomes, ER, or Golgi apparatus rather than nuclei, mitochondria, or the cytosol.
MHV replicase and structural proteins segregate into distinct cellular membrane populations.
We next determined if the cellular membrane populations in P100 that contained the viral proteins could be further separated. We chose iodixanol as our gradient medium since it allows the fractionation of membranes under isotonic conditions. The P100 pellet was resuspended and loaded onto a continuous iodixanol gradient, and following centrifugation, 10 fractions were collected and assayed for the presence of MHV structural and gene 1 proteins. Similar results were obtained when the experiments were repeated using sucrose gradients (data not shown).
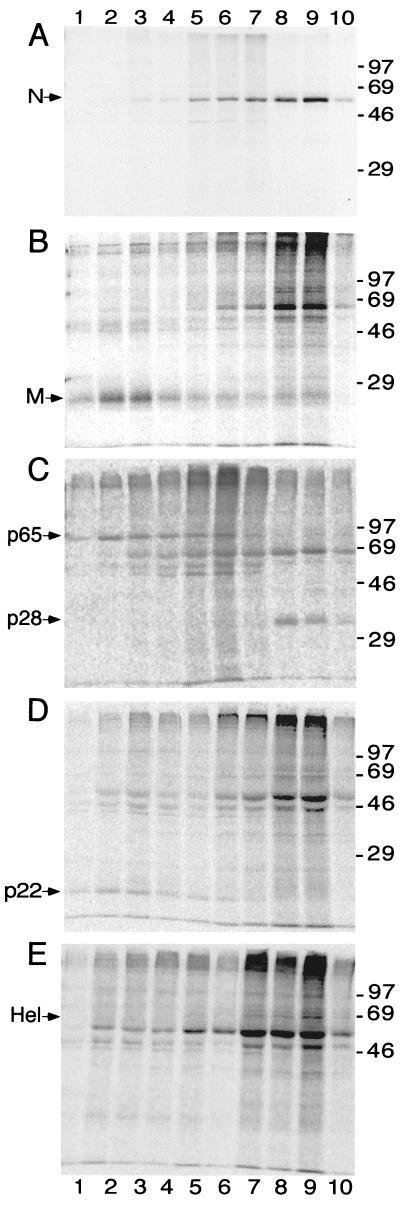
N was detected with cellular membranes in fractions 6 through 9 (1.09 to 1.13 g/ml) with a distinct peak of detection in fractions 8 and 9 (1.12 to 1.13 g/ml) (Fig. 3A). (Fraction 1 was the “top,” least dense fraction.) In contrast, M was concentrated toward the top of the gradient, associated with relatively less dense membranes in fractions 2 and 3 (1.05 to 1.06 g/ml) (Fig. 3B). Although small amounts of M were detected in almost all fractions, M was concentrated in less dense membranes. Thus, the peak concentrations of N and M were at opposite ends of the gradient, demonstrating that these two structural proteins were predominantly associated with distinct membrane densities at 7 h p.i.
FIG. 3.
Iodixanol gradient fractionation of P100 pellet and immunoprecipitation of gene 1 and structural proteins. The P100 pellet from MHV-infected cells was resuspended and fractionated on the iodixanol gradient as described in Materials and Methods. Gradient fractions were immunoprecipitated, analyzed by SDS–12% PAGE, and subjected to fluorography (A) or phosphoimager analysis (B, C, D, and E). Gels are aligned so that labels at top and bottom apply to all gels. Lanes 1 indicate the top of the gradient (less dense), and lanes 10 indicate the bottom of the gradient (more dense). Marker proteins (molecular masses in kilodaltons) are to the right of the gels, and proteins of interest are to the left of the gels. (A) anti-N; (B) anti-M; (C) UP102, anti-p28/p65; (D) B4, anti-p1a-22; (E) B1, anti-Hel.
We next examined the distribution of the gene 1 proteins in the gradient. Hel and p28 were detected exclusively in fractions 8 and 9, the same fractions containing peak concentrations of N (Fig. 3C and E). The UP102 antibody that detected p28 also precipitated p65, the protein immediately adjacent to p28 in the polyprotein (Fig. 1) (14). However, in contrast to the localization of p28, p65 was detected in fractions 2 through 6 (1.05 to 1.09 g/ml) (Fig. 3C), demonstrating no overlap of p65-containing membranes with the membrane populations containing p28. The complete separation of p28 and p65 indicated that, once cleaved, these proteins segregated to different membranes. The experiment also was consistent with our previous studies showing that antibodies directed against p28 did not result in coprecipitation of p65 (16). The p1a-22 protein had the same fractionation pattern as p65, with concentration in fractions 2 through 6 and no overlap with p28 or Hel (Fig. 3D). Together, the fractionation results demonstrated that at least two distinct populations of MHV protein-associated membranes were present during infection and that these populations were readily separable by density gradient fractionation under native (nondetergent, nonreducing) conditions.
MHV RNA localizes to the membrane population containing p28, Hel, and N.
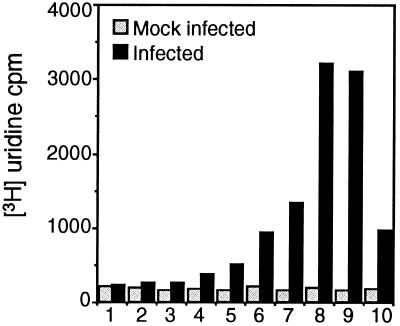
Since the gene 1 proteins segregated into at least two populations of membrane-protein complexes, we sought to determine if viral RNA was localized to one or both of the membrane populations. Mock-infected and infected cells were metabolically labeled with [3H]uridine in the presence of actinomycin D, and cell lysates were subjected to differential centrifugation followed by gradient fractionation of P100. Actinomycin D-resistant, TCA-precipitable [3H]uridine incorporation was measured for each fraction. No actinomycin D-resistant [3H]uridine incorporation was detected in mock-infected cells compared to background either in crude membrane pellets or in any gradient fraction. In MHV-infected cells, a small amount of the actinomycin D-resistant viral RNA was detected in the nuclear (P1) and intermediate-speed (P2.3) pellets, as well as the S100 cytosol, but the majority of the viral RNA synthesis was detected in P100, the fraction that also contained all gene 1 proteins. When P100 was fractionated, a single distinct peak of [3H]uridine incorporation was seen in fractions 8 and 9, the same fractions containing N, Hel, and p28 (Fig. 4). Smaller amounts of viral RNA were detected in fractions 6, 7, and 10, with none detected in fractions 1 to 5. The detection of viral RNA in only the membranes containing p28, Hel, and N after a 2-h labeling period (6 to 8 h p.i.) demonstrated that viral RNA was retained in the dense membrane population.
FIG. 4.
Detection of new viral RNA in iodixanol gradient fractions of P100. MHV-infected (black bars) or mock-infected (gray bars) DBT cells were labeled with [3H]uridine in the presence of actinomycin D, lysed, and subjected to differential centrifugation, and the P100 pellet was fractionated on an iodixanol gradient. Equal amounts of the gradient fractions were precipitated with 8% TCA, spotted onto Whatman glass filters, and counted by scintillation counter. TCA counts are shown to the left. Numbers 1 through 10 indicate fractions as in Fig. 3.
Identification of cellular membranes in iodixanol gradient fractions.
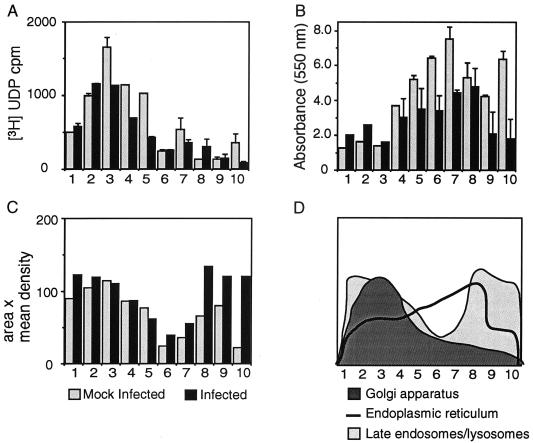
To identify the intracellular membranes contained within the iodixanol gradient fractions, the gradient fractions of P100 were assayed for galactosyltransferase (Golgi apparatus) and NADPH-cytochrome c reductase activity (ER) (23) as well as for the protein LAMP-1 (late endosomes and lysosomes) (12). The distributions of Golgi enzymes were similar between mock-infected and infected cells, with galactosyltransferase activity detected in fractions 2 through 5 in both mock-infected and MHV-infected DBT cells (Fig. 5A). The cosegregation of M with Golgi membranes was consistent with a number of studies showing concentration of M on Golgi membranes and provided a positive control for cosegregation of viral proteins with organelle membranes (26–28, 38, 39).
FIG. 5.
Determination of cell organelle marker proteins in the iodixanol gradient fractions of P100. Numbers underneath each figure are gradient fractions as in Fig. 3. Black bars in panels A, B, and C indicate MHV-infected cells, whereas gray bars indicate mock-infected cells. (A) Assay for Golgi apparatus. Equal volumes of gradient fractions were incubated with ovalbumin and [3H]UDP-galactose to test for the presence of galactosyltransferase. (B) Assay for ER. Samples were incubated with NADPH and cytochrome c, and measurements were taken at 550 nm for 10 min to assess NADPH-cytochrome c reductase activity. (C) Assay for endosomes and lysosomes. Equal volumes of each fraction were separated by SDS–12% PAGE, transferred, and subjected to Western blot analysis with LAMP-1 antisera. Results were analyzed by densitometry (area in pixels × mean density of pixels). (D) Summary and comparison of infected cell results from panels A to C. For panels A and B, each sample was standardized to the protein concentration of the infected sample.
Assays for NADPH-cytochrome c reductase activity demonstrated that ER membranes were more broadly distributed across the center of the gradient in fractions 4 through 8, with little ER enzymatic activity in fractions 1 through 3 or 9 and 10 (Fig. 5B). Iodixanol efficiently separates the ER from lysosomes because the ER has a lower density in iodixanol than in other gradient matrices (22). There was little to no overlap of ER membranes with the fractions containing M, p1a-22, or p65. There was, however, significant overlap between the distribution of ER membranes on the gradient and that of those containing N, Hel, p28, and the viral RNA.
When the iodixanol gradient fractions were analyzed for the presence of LAMP-1, two distinct populations of LAMP-1-containing membranes were detected in fractions 1 through 3 and fractions 8 through 10, respectively (Fig. 5C). The distribution of LAMP-1 on the gradient mirrored the bimodal distribution of membranes containing gene 1 proteins. Previous studies of organelle separation on iodixanol gradients have demonstrated a clear separation of endosomes and lysosomes to the top (less dense) and bottom (more dense) fractions, respectively, similar to the pattern observed in our study (22). The same bimodal pattern of LAMP-1 was seen in both mock-infected and MHV-infected cells, indicating that, even at 7 h p.i., the basic character of LAMP-1-positive membranes was retained.
The cofractionation of LAMP-1 and galactosyltransferase activity in fractions 1 through 5 provided an answer for why p1a-22 was detected in fractions that contained M. Specifically, it appeared that Golgi membranes containing M had the same density in iodixanol as LAMP-1-positive membranes containing p1a-22. Together, the cell marker experiments provided biochemical confirmation of the origin of gene 1 protein-containing complexes from LAMP-1-positive organelles and also suggested a role for membranes of the ER in MHV replication. They also confirmed that, at 7 h p.i., viral RNA and the associated viral proteins p28, Hel, and N were not associated with membranes of the Golgi apparatus.
p65 colocalizes with p1a-22.
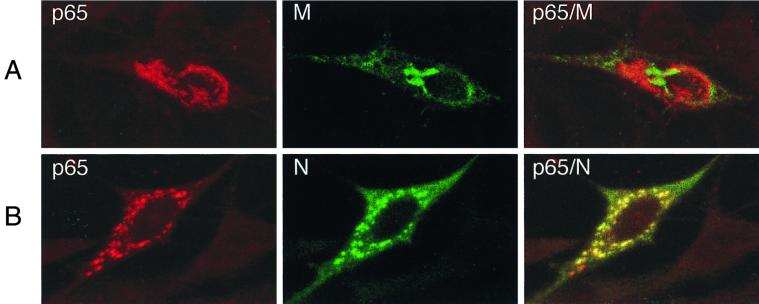
The gene 1 proteins p1a-22 and p65 were detected in fractions that also contained the structural M protein, a protein known to localize to Golgi membranes. Since the localization of p65 in MHV-infected cells had not been determined using specific antibodies, either biochemically or by imaging studies, we investigated whether p65 might be localizing with M in Golgi membranes or localizing to a unique population of membranes. When MHV-infected cells were probed for both M and p65 using laser confocal microscopy, it was immediately apparent that M and p65 localized to different membrane populations (Fig. 6A). In contrast to the localization of M, p65 localized to discrete, punctate structures throughout the cytoplasm. When cells were dual labeled for N and p65, p65 colocalized almost entirely with N (Fig. 6B), a result identical to that previously obtained with p1a-22 and N (9). The results indicated that both p65 and p1a-22 localized to membranes that were distinct from the Golgi. Additionally, they appeared to colocalize with N by confocal IF but were distinct from N upon cell fractionation. Thus, it appeared that p65- and p1a-22-containing membranes were in such close approximation to membranes containing N as to be indistinguishable by light microscopy (<200 nm with the 40×, 1.3-numerical-aperture lens used).
FIG. 6.
Subcellular localization of p65 and MHV structural proteins in DBT cells. Infected cells on glass coverslips were fixed at 5.5 h p.i. and prepared for IF using primary antibodies against p65, N, and M, followed by Cy2 (green)- or Cy3 (red)-conjugated anti-rabbit (p65) or anti-mouse (N and M) secondary antibodies. Confocal images were obtained on a Zeiss LSM 410 confocal microscope. Images were merged using Adobe Photoshop 5.0.
DISCUSSION
Gene 1 proteins segregate to distinct but closely associated membrane complexes.
Biochemical fractionation experiments revealed that “groups” of gene 1 proteins target different populations of intracellular membranes. The experiments were performed in isotonic media in the absence of reducing agents or detergent in order to retain membrane integrity and protein interactions. The complete separation of the membrane populations on the gradient thus strongly suggested that the gene 1 protein-containing membranes are not fused and are not extensions of the same membranes. By all analyses, the two gene 1 protein-containing membrane populations were distinct. The gradient separation of gene 1 proteins such as p1a-22 and Hel to distinct membranes appeared to differ from confocal IF and electron microscopy studies in which the same proteins colocalized to varying degrees (9, 40). In fact, the present study provides a biochemical explanation for the observations made by cell imaging approaches. While the study of van der Meer et al. showed labeling for Hel, Pol, and p1a-22 together with viral RNA in membranes containing markers for the endosomal pathway, the immuno-EM analysis also revealed several different morphologies for the gene 1 protein-containing membranes, and in addition to areas where Pol and p1a-22 colabeled membranes, there were also distinct areas where Pol or p1a-22 was detected in the absence of other gene 1 proteins (40). In a more recent study, Bost et al. showed by confocal microscopy that, while p1a-22 substantially overlapped with Hel and N, p1a-22 also was present in foci distinct from the other gene 1 proteins (9). In addition, the membranes containing p1a-22 appeared to tightly interdigitate with, but remain distinct from, membranes containing N and Hel. The gradient fractionation results demonstrate that, while the membranes containing p1a-22 are distinct from those containing N, Hel, and viral RNA (and presumably Pol), they may be so closely associated or interdigitated that they may constitute a single multipartite complex of membranes that cannot always be resolved by light or electron microscopic approaches. The ease of separation of the membranes also suggests that any interactions between the membrane populations are not the result of fusion, proteins that span both membranes, disulfide bridges, or strong hydrophobic forces.
MHV RNA synthesis is associated with one population of gene 1 protein-containing membranes.
It has been known for many years that coronavirus RNA synthesis occurs in association with intracellular membranes (18), and recent studies of the coronavirus transmissible gastroenteritis virus (TGEV) have demonstrated that viral RNA can be identified in membrane fractions from lysates of whole cells separated on cesium chloride gradients (36). Our study is the first to biochemically demonstrate association of viral RNA with a single population of membranes containing gene 1 proteins and N. Our results differed from those of Sethna and Brian (36) by showing a single peak of viral RNA in the gradient fractions from MHV-infected cells, whereas more than one peak of viral RNA was detected in TGEV-infected cells. Because of this difference, we repeated the experiment but separated the gradient into 50 fractions instead of 10, in order to eliminate “averaging” across fractions and to detect minor peaks that might be lost in larger fractions. When this was done, the single peak of viral RNA was even more pronounced and sharply focused in the same area of the gradient, and no other minor peaks of [3H]uridine incorporation were detected (data not shown).
The apparent differences are likely due to the conditions used to perform the study. Sethna and Brian fractionated lysates of whole cells, whereas our study used gradient fractions from a high-speed, small-membrane pellet (P100) containing the gene 1 proteins. Thus, the TGEV study may have identified populations of viral RNA that would not have pelleted with P100 in our studies. In that regard, it is important to note that, although we detected the majority of RNA in the P100 pellet, a smaller amount of [3H]uridine incorporation was detected in the nuclear (P1) and intermediate (P2.3) pellets and in the S100 cytosol (data not shown). Sethna and Brian also recognized that the high salt content of the CsCl2 might cause disruption of membranes and alterations in the distribution of RNA. In contrast, the iodixanol gradients used in our study were iso-osmolar and nondisruptive to membrane and protein interactions (20), and so it is likely that our results reflect native conditions.
Our results demonstrated that viral RNA predominantly concentrates in small membranes that also contain gene 1 proteins. In addition, the remarkable segregation of viral RNA to a single population of membranes in association with specific gene 1 and structural proteins strongly suggests that these membranes are the active replication complexes. The coronaviruses use a complex strategy of genomic and subgenomic RNA transcription and replication and target proteins to different compartments. It is therefore certainly possible that viral RNA is present in sites other than replication complexes. It will be important to define the RNA species present in crude cell fractions as well as in gradient fractions to determine if the virus performs all of its RNA synthetic activities in the gradient-purified membranes.
Hel, p28, and N are associated with sites of viral RNA synthesis while p65 and p1a-22 are not.
The experiments in this report confirmed our previous conclusions that N and Hel were intimately associated with sites of viral RNA synthesis and support the prediction that N and Hel are directly involved in the processes of replication and transcription (17, 30). More interesting perhaps was the association of p28 with the membranes containing Hel and N. p28 was the first gene 1 cleavage product identified in vitro and in virus-infected cells (13, 16), but its role in coronavirus replication remains unknown. Based on its calculated pI of 9.4, p28 has been predicted to be an RNA binding protein (13). Nonconfocal IF studies of p28 detected a diffuse staining pattern throughout the cytoplasm (4). Interestingly, p28 was the only gene 1 protein among those tested that was also seen in the S100 cytosol in addition to the P100 pellet, which may account for the previous IF findings. However, the majority of p28 was detected in the P100 pellet, and when fractionated, it localized only to fractions containing Hel. Its cofractionation with Hel and viral RNA suggests that it plays a direct role in viral RNA synthesis or in the formation or maintenance of complexes where viral RNA synthesis occurs.
Equally intriguing was the isolation of a discrete, less dense population of membranes that contained two previously identified gene 1 proteins, p65 and p1a-22 (14, 32) (Fig. 3C and D). p1a-22 is one of four small proteins that comprise the carboxy-terminal 59 kDa of pp1a and is likely one of the first proteins cleaved by 3CLpro. There are no motifs that predict functions of p1a-22, but the four proteins that include p1a-22 are conserved among coronaviruses in their location and relative size. The separation of p1a-22 from Hel indicates that it is less likely to be involved directly in viral RNA synthesis. The segregation of p65 to membranes distinct from those containing p28 was interesting because these two proteins are juxtaposed in the polyprotein and are cleaved by the first translated viral proteinase, PLP-1 (15, 19). Like p28, p65 is unique to MHV, but its function during virus replication remains unknown. Their complete separation by density gradient fractionation suggests that one or both of these proteins target distinct intracellular membranes following translation and processing.
Of mice and membranes.
Biochemical fractionation of infected cell lysates confirmed that both populations of membranes containing gene 1 proteins possessed markers for membranes in the endosomal-lysosomal pathways and also confirmed that Golgi membranes are not present in complexes active in viral RNA synthesis. However, our study also raises important questions concerning the origin of the specific membranes in the replication complexes and the mechanism of replication complex formation. It was of interest that the membranous compartments of virus-infected cells retained their basic characteristics even at later times of infection when substantial syncytium formation was present. Thus, based on the known density of cellular membranes in iodixanol and the similarity in fractionation patterns between mock-infected and infected cells, it would appear that membranes containing p28, Hel, N, and RNA are more consistent with lysosomes whereas those containing p1a-22 and p65 are more consistent with endosomes (22); however, this conclusion would be at odds with immuno-EM observations and with our own studies suggesting that gene 1 proteins do not target lysosomes (data not shown). It is possible that the replication complexes occur on membranes at the transition between endosomes and lysosomes and that this might account for the observed density of the membranes. Alternatively, it is possible that incorporation of proteins and especially viral RNA may alter the density of endosomal membranes so that they cofractionate with lysosomes. Our results also suggest a role for membranes of the ER during MHV RNA synthesis due to the significant overlap of the fractions containing p28, Hel, N, and viral RNA with those containing ER-derived NADPH-cytochrome c reductase activity. Certainly, translation of gene 1 from the input genome RNA is required to initiate replication, but whether the ribosomes are derived from ER or from free polysomes requires further investigation. Finally, it is not clear how the different gene 1 proteins target specifically one membrane population. We would speculate that the membranes containing RNA would be the most likely initial site of gene 1 protein translation and processing and therefore that proteins such as p1a-22 and p65 would need to target the other membrane population. How and why they do this remain to be determined, and the answers to these questions will likely reveal important mechanisms by which the coronaviruses use the cell to mediate viral replication.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI-26603 (M.R.D.) and AI-01479 (M.R.D.).
We thank Hal Love and Patricia Anderson for their assistance. We acknowledge critical reading of the manuscript by Anne Gibson Bost, the technical assistance of Xiaotao Lu, the intellectual and physical strength of Erik Prentice, and the assistance of David Piston and Jonathon Sheehan in the Molecular Imaging Shared Resource of the Vanderbilt University Cancer Center (IP30CA68485). We also thank John Fleming at the University of Wisconsin for kindly providing the anti-M (J.1.3) and anti-N (J.3.3) antibodies.
REFERENCES
- 1.Anderson R, Wong F. Membrane and phospholipid binding by murine coronaviral nucleocapsid N protein. Virology. 1993;194:224–232. doi: 10.1006/viro.1993.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch W E, Dunphy W G, Braell W A, Rothman J E. Reconstitution of the transport of proteins between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 1984;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- 3.Balch W E, Rothman J E. Characterization of protein transport between successive compartments of the Golgi apparatus: asymmetric properties of donor and acceptor activities in a cell-free system. Arch Biochem Biophys. 1985;240:413–425. doi: 10.1016/0003-9861(85)90046-3. [DOI] [PubMed] [Google Scholar]
- 4.Bi W, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Intracellular localization of polypeptides encoded in mouse hepatitis virus open reading frame 1A. Adv Exp Med Biol. 1995;380:251–258. doi: 10.1007/978-1-4615-1899-0_40. [DOI] [PubMed] [Google Scholar]
- 5.Bi W, Pinon J D, Hughes S, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Localization of mouse hepatitis virus open reading frame 1a derived proteins. J Neurovirol. 1998;4:594–605. doi: 10.3109/13550289809114226. [DOI] [PubMed] [Google Scholar]
- 6.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla P J, Hughes S A, Pinon J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 8.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bost A G, Carnahan R H, Lu X T, Denison M R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J Virol. 2000;74:3399–3403. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breedenbeek P J, Pachuk C J, Noten A F H, Charite J, Luytjes W, Weiss S R, Spaan W J M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brierley I, Boursnell M E G, Binns M M, Billimoria B, Blok V C, Brown T D K, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J W, Murphy T L, Willingham M C, August J T. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison M, Perlman S. Translation and processing of MHV-A59 virion RNA in reticulocyte lysates and infected cells. Adv Exp Med Biol. 1987;218:155–156. doi: 10.1007/978-1-4684-1280-2_18. [DOI] [PubMed] [Google Scholar]
- 14.Denison M R, Hughes S A, Weiss S R. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology. 1995;207:316–320. doi: 10.1006/viro.1995.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison M R, Kim J C, Ross T. Inhibition of coronavirus MHV-A59 replication by proteinase inhibitors. Adv Exp Med Biol. 1995;380:391–397. doi: 10.1007/978-1-4615-1899-0_64. [DOI] [PubMed] [Google Scholar]
- 16.Denison M R, Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol. 1986;60:12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denison M R, Spaan J M, van der Meer Y, Gibson C A, Sims A C, Prentice E, Lu X T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis D E, Brian D A. RNA-dependent RNA polymerase activity in coronavirus-infected cells. J Virol. 1982;42:153–164. doi: 10.1128/jvi.42.1.153-164.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong S, Baker S C. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford T, Graham J, Rickwood D. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal Biochem. 1994;220:360–366. doi: 10.1006/abio.1994.1350. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya A E, Koonin E V, Lai M M-C. Putative papain-related thiol proteases of positive-strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham J, Ford T, Rickwood D. The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal Biochem. 1994;220:367–373. doi: 10.1006/abio.1994.1351. [DOI] [PubMed] [Google Scholar]
- 23.Graham J M. Identification of subcellular fractions from mammalian cells. In: Graham J M, Higgins J A, editors. Biomembrane protocols I. Isolation and analysis. Vol. 19. Totowa, N.J: Humana Press; 1993. pp. 1–20. [Google Scholar]
- 24.Hirano N, Fujiwara K, Matumoto M. Mouse hepatitis virus (MHV-2); plaque assay and propagation in mouse cell line DBT cells. Jpn J Microbiol. 1976;20:219–225. [PubMed] [Google Scholar]
- 25.Kim J C, Spence R A, Currier P F, Lu X T, Denison M R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krijnse-Locker J, Ericsson M, Rottier P J, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krijnse-Locker J, Klumperman J, Oorschot V, Horzinek M, Geuze H, Rottier P. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J Biol Chem. 1994;269:28263–28269. [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, LaMonica N, Tuler J, Bagdzhadhzyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu X T, Lu Y Q, Denison M R. Intracellular and in vitro translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology. 1996;222:375–382. doi: 10.1006/viro.1996.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X T, Sims A C, Denison M R. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J Virol. 1998;72:2265–2271. doi: 10.1128/jvi.72.3.2265-2271.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y Q, Denison M R. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology. 1997;230:335–342. doi: 10.1006/viro.1997.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pachuk C J, Breedenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi S T, Schiller J J, Kanjanahaluethai A, Baker S, Oh J, Lai M M C. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac- cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 39.Tooze J, Tooze S A. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur J Cell Biol. 1985;37:203–212. [PubMed] [Google Scholar]
- 40.van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J M, Krijnse Locker J. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]