Abstract
The small envelope (E) protein has recently been shown to play an essential role in the assembly of coronaviruses. Expression studies revealed that for formation of the viral envelope, actually only the E protein and the membrane (M) protein are required. Since little is known about this generally low-abundance virion component, we have characterized the E protein of mouse hepatitis virus strain A59 (MHV-A59), an 83-residue polypeptide. Using an antiserum to the hydrophilic carboxy terminus of this otherwise hydrophobic protein, we found that the E protein was synthesized in infected cells with similar kinetics as the other viral structural proteins. The protein appeared to be quite stable both during infection and when expressed individually using a vaccinia virus expression system. Consistent with the lack of a predicted cleavage site, the protein was found to become integrated in membranes without involvement of a cleaved signal peptide, nor were any other modifications of the polypeptide observed. Immunofluorescence analysis of cells expressing the E protein demonstrated that the hydrophilic tail is exposed on the cytoplasmic side. Accordingly, this domain of the protein could not be detected on the outside of virions but appeared to be inside, where it was protected from proteolytic degradation. The results lead to a topological model in which the polypeptide is buried within the membrane, spanning the lipid bilayer once, possibly twice, and exposing only its carboxy-terminal domain. Finally, electron microscopic studies demonstrated that expression of the E protein in cells induced the formation of characteristic membrane structures also observed in MHV-A59-infected cells, apparently consisting of masses of tubular, smooth, convoluted membranes. As judged by their colabeling with antibodies to E and to Rab-1, a marker for the intermediate compartment and endoplasmic reticulum, the E protein accumulates in and induces curvature into these pre-Golgi membranes where coronaviruses have been shown earlier to assemble by budding.
Coronaviruses, a family of viruses belonging to the newly established order of the Nidovirales (for reviews, see references 8 and 37) have enveloped virions containing a nonsegmented, plus-stranded RNA genome. The RNA is packaged by the nucleocapsid (N) protein into a helical nucleocapsid. The surrounding envelope contains three, and sometimes four, membrane proteins. The spike (S) protein, a type I glycoprotein, occurs as trimers that constitute the characteristic surface projections. These function primarily in virus entry, being responsible for binding to the receptor on the target cell and for mediating fusion of viral and cellular membranes. The membrane (M) protein is a triple-spanning glycoprotein. It is the most abundant envelope protein component having essential functions in virus assembly. The hemagglutinin-esterase protein is present in only a subset of coronaviruses. The type I glycoprotein occurs in virions in disulfide-linked homodimeric form. Its biological role in the virus life cycle has not been well established.
The small envelope (E) protein was only recently recognized as a structural component of the coronavirion (12, 26, 48, 49). Although very little is still known about its features, the E protein appears to be surprisingly important for assembly of the viral envelope. By coexpression of the genes encoding the mouse hepatitis virus strain A59 (MHV-A59) membrane proteins we showed that virus-like particles (VLPs) morphologically mimicking normal virions were produced only when the E protein was present, while the S protein was dispensable (48). Similar particles were observed after coexpression of transmissible gastroenteritis virus (TGEV) membrane proteins (1a).
Coronavirus E proteins vary in size from about 76 to 109 amino acids (for a review, see reference 38). Consistent with their membrane association (12, 40, 48, 49), the proteins are generally quite hydrophobic in nature, particularly in their N-terminal half (see Fig. 1). The MHV-A59 E protein was reported to be acylated on the basis of a biochemical assay (49), but attempts to directly label the TGEV protein with palmitic acid failed (12). By immunofluorescence, the E protein was observed in infected cells in a granular (IBV) (40) or punctate (BCV and MHV-A59) (1, 49) pattern, as well as at the plasma membrane (12, 40, 49). The cell surface staining with a C-terminus-specific antibody led Godet et al. (12) to suggest a CexoNendo membrane topology for the TGEV E protein, i.e., with its C and N termini exposed luminally and cytoplasmically, respectively.
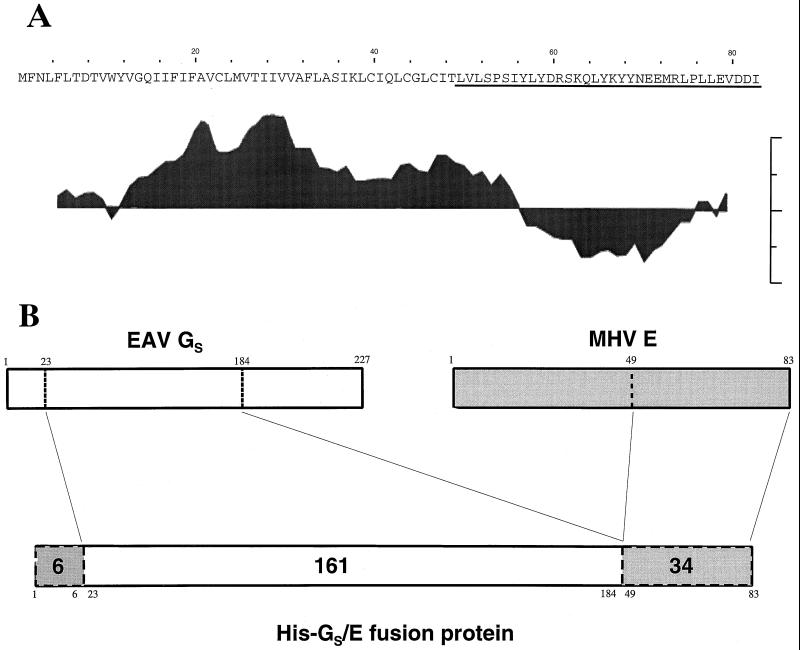
FIG. 1.
(A) Amino acid sequence of the MHV-A59 E protein and its hydropathy profile as determined by Kyte and Doolittle (23) with a 8-residue moving window. Peaks extending upwards indicate hydrophobic domains; those pointing downwards represent hydrophilic regions. (B) Structure of the fusion protein for antibody preparation against E protein. A construct was prepared consisting of the ectodomain of the EAV GS protein (residues 23 to 184) and the MHV-A59 E protein (residues 49 to 83, underlined in panel A) preceded by a 6-histidine stretch.
Very recently Fischer et al. (10) described the effects of mutations in the E protein of MHV-A59. Using clustered charged-to-alanine mutagenesis and targeted RNA recombination, two mutant viruses were obtained that were partially temperature sensitive, forming small plaques at the nonpermissive temperature, and markedly thermolabile when grown at the permissive temperature. Most interestingly, one of these viruses appeared to have strikingly aberrant morphology when viewed by electron microscopy. Many virions showed pinched and elongated shapes, which is consistent with a role for the E protein in particle morphogenesis.
In order to obtain more insight into the function of the E protein in coronavirus infection, particularly regarding its role in viral assembly, we have analyzed a number of its basic features. We have studied, in addition to its appearance and fate in infected cells, its independent properties by expression, as well as its topology in cellular and viral membranes.
MATERIALS AND METHODS
Cells and viruses.
Mouse L cells, OST7-1 cells (9), and RK13 cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS)–100 IU of penicillin per ml–100 μg of streptomycin per ml (DMEM–10% FCS), supplemented in the case of OST7-1 cells with 400 μg of G-418 (Geneticin; Gibco BRL) per ml. BHK-21 cells and CEF cells were maintained in Glasgow minimal essential medium (MEM) containing the same additional ingredients.
MHV-A59 was propagated in Sac(−) cells as described previously (42). The recombinant vaccinia viruses vTF7-3 and MVA-T7 expressing bacteriophage T7 RNA polymerase were propagated in RK13 and CEF cells, respectively, as described previously (11, 43).
Expression vectors.
The expression construct pT7Ts-E carrying the MHV-A59 E gene behind a T7 promoter was made by using the vector pT7Ts (a kind gift of P. Krieg). The E gene was obtained as a PCR fragment amplified from cDNA clone pRG86 (2) by using primer 471 (5′-dCACGCAGCTCGAAACATATGTTTA-3′) and primer 496 (5′-dGGATTAGATATCATCCACCTCTA-3′). It was cloned into pNoTa (5 Prime→3 Prime, Inc.). From the resulting plasmid pNoTa/T7-5b, the E gene was subsequently retrieved with NdeI and BamHI, and this fragment was treated with Klenow DNA polymerase. pT7Ts was first digested with EcoRI and XbaI and then treated with Klenow enzyme and religated. The plasmid was then cut with SpeI and treated again with Klenow DNA polymerase, and finally the E gene fragment was ligated into it to yield pT7Ts-E.
The expression plasmid pTUM-M containing the MHV-A59 M gene behind a T7 promoter has been described elsewhere (28), as has pAVI02, a pBluescript KS(+) construct carrying the equine arteritis virus (EAV) Gs gene (6).
Antibodies.
To obtain a serum against the MHV-A59 E protein, we constructed a plasmid encoding an N-terminally His-tagged fusion protein of the major part of the EAV Gs protein ectodomain linked to the C-terminal 34-residues sequence of the E protein (Fig. 1B). To this end, we amplified the relevant region of the plasmid pNoTA/T7-5b by PCR using the primers 485 (5′-dGGAATTCTTTGGTGCTGTCCCCTTC-3′; nucleotides 15554 to 15571 of the MHV-A59 RNA sequence) and the M13 reverse sequence primer (Pharmacia). This PCR product was digested with EcoRI and HindIII and cloned into pBluescript SK(−) to generate pBM.cE. To obtain the fragment encoding the 161-residues Gs polypeptide, we performed a PCR with the primers 483 (5′-dCGGGATCCATCGCCCGCAGCTTGG-3′, nucleotides 9897 to 9914 of the EAV RNA sequence) and primer 484 (5′-dGGAATTCAACACTTCCACAGATGAAGC-3′; genomic positions 10362 to 10382). This fragment was cloned as a BamHI/EcoRI fragment into pBluescript SK(−) to generate pBE.GS-ecto. Then the EcoRI/HindIII fragment from pBM.cE was cloned into pBE.GS-ecto behind the EAV fragment which resulted in the unintentional insertion of two codons at the Gs-E junction site adding two amino acids (Asn and Ser). The chimeric Gs-E fragment was cloned as a BamHI fragment into the His-tag vector pQE10 (Qiagen) yielding pGSectoEendo. The protein was expressed in Escherichia coli, isolated, and purified on a Ni-nitriloacetic acid column (Invitrogen) according to the manufacturer's instructions. Antibodies were elicited by subcutaneous injection of the fusion protein (75 μg in Freund complete adjuvant) into a rabbit from which blood had been taken the day before to obtain a preserum (designated preE). The rabbit was boosted similarly after 3, 7, 11, and 15 weeks using 250 μg of protein in Freund incomplete adjuvant each time. Animals were bled 17 weeks after the first immunization.
The monoclonal antibody J1.3 (a kind gift of J. Flemming) recognizes the N-terminal domain of the MHV-A59 M protein (designated αMN), as has been described (5). Also described previously was a rabbit serum against a peptide corresponding to the M protein's C terminus (αMC [19rsqb;), as well as a polyclonal rabbit serum against MHV virions (αMHV [32rsqb;).
Infection, transfection, and metabolic labeling.
Subconfluent monolayers of OST7-1 cells grown in 10-cm2 dishes were washed with phosphate-buffered saline (PBS) containing 50 μg of DEAE-dextran per ml (PBS-DEAE) and 1% of FCS and inoculated using this medium with MHV-A59 at a multiplicity of infection (MOI) of 10 to 50 for 60 min at 37°C.
For expression of the MHV-A59 M and E genes and of the EAV Gs gene, subconfluent monolayers of OST7-1 or BHK-21 cells in 10-cm2 dishes were washed with DMEM and inoculated with vTF7-3 or MVA-T7 at an MOI of 10 in DMEM for 45 min at 37°C. Cells were then washed with DMEM and transfected with the plasmid of choice. For this purpose, 200 μl of DMEM containing 10 μl of Lipofectin reagent (Life Technologies, Inc.) was mixed with 5 μg of plasmid DNA and added to the cells. After a 10-min incubation at room temperature, 800 μl of DMEM was added, and the cells were incubated further initially at 37°C for 1 h and subsequently at 32°C for the indicated times; in the case of MVA-T7-driven expression the cells were kept at 37°C at all times.
For radioactive labeling of proteins, MHV-A59-infected cells and cells expressing genes from plasmids were starved for 30 min by incubation with MEM lacking methionine and cysteine (GIBCO) and supplemented with 10 mM HEPES (pH 7.4). Cells were labeled with 80 μCi of 35S In Vitro Labeling Mix (Amersham) in the same medium for the times indicated. For pulse-chase experiments, the cells were washed twice after the labeling with prewarmed chase medium (DMEM–10% FCS, 2 mM methionine and cysteine, and 10 mM HEPES [pH 7.4]) and incubated further in the same medium.
Immunoprecipitation, immunoisolation, and protein analysis.
After labeling, cells were washed once with PBS containing 50 mM Ca2+ and 50 mM Mg2+. They were then lysed on ice in 1 ml of lysis buffer (20 mM Tris, pH 7.6; 150 mM NaCl; 1% Nonidet P-40 [NP-40]; 0.5% deoxycholate; and 0.1% sodium dodecyl sulfate [SDS]) per 10-cm2 dish. The lysates were cleared by centrifugation for 15 min at 13,000 rpm at 4°C in an Eppendorf centrifuge. To analyze proteins present in the culture media, cell supernatants were taken off, cleared by centrifugation for 15 min at 4°C and 4,000 rpm, and then mixed with a one-fifth volume of a 5× concentrated lysis buffer: 100 mM Tris (pH 7.6), 150 mM NaCl, 5% (vol/vol) NP-40, 2.5% (wt/vol) deoxycholate (DOC), 0.5% (wt/vol) SDS. To analyze proteins present in cells and culture media together, as was done in the experiments of Fig. 2 and 3, total lysates were prepared by adding a one-fifth volume of 5× concentrated lysis buffer to the culture media. The combined lysates of cells and media were cleared by centrifugation as described above for conventional cell lysates.
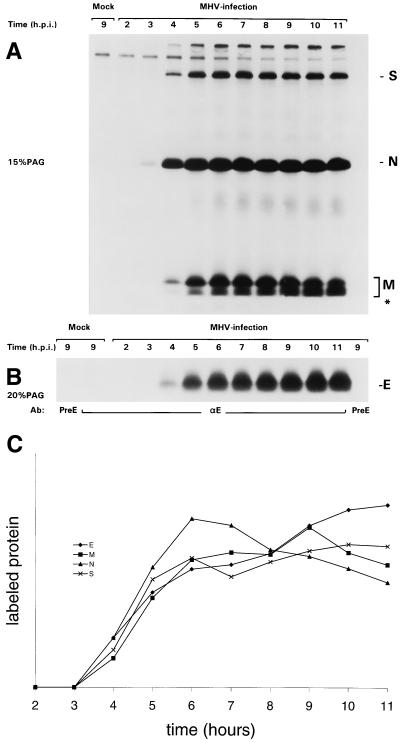
FIG. 2.
Kinetics of appearance of the E protein in MHV-A59-infected cells. MHV-infected and mock-infected OST7-1 cells were labeled with 35S-labeled amino acids (80 μCi/10-cm2 dish) for different 1-h periods starting at the indicated times after infection. Combined lysates of cells and culture media were then prepared, and immunoprecipitations were carried out with different antisera. (A) Proteins precipitated with the polyclonal anti-MHV serum were analyzed in 15% PAG. (B) Proteins precipitated by the anti-E (αE) or the preE serum were analyzed in 15% PAG. Radioactivities in the bands representing the different viral proteins were quantitated, taking for M all the different forms, including the lower band indicated by an asterisk. The results are compiled in panel C. They were normalized by placing the added total of all measurements for each protein at 100 and expressing each measurement as the fraction of this total.
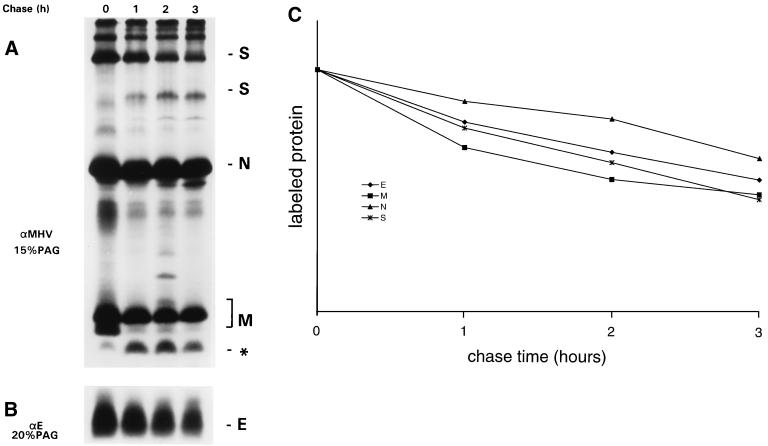
FIG. 3.
Stability of the E protein in MHV-A59 infection. MHV-infected OST7-1 cells were labeled with 35S-labeled amino acids (80 μCi/10-cm2 dish) for 30 min starting at 6 hpi. Combined lysates of cells and culture media were prepared either immediately or after different chase periods as indicated. Viral proteins were immunoprecipitated with polyclonal anti-MHV serum (αMHV) or anti-E (αE) serum and analyzed in 15% (A) and 20% (B) PAG, respectively. The radioactivities in the viral proteins were quantitated, including for S protein also the cleaved form of the protein and for M the faster-migrating band indicated by an asterisk (C). The results are compiled in panel C. For each protein the radioactivity in the chase samples was related to that observed after the pulse labeling, which was set at 100.
For immunoprecipitation of proteins, 100- or 150-μl aliquots of lysates were diluted to 1 ml with immunoprecipitation buffer (IP buffer; 20 mM Tris [pH 7.6], 150 mM NaCl, 5 mM EDTA, 0.5% [vol/vol] NP-40, 0.1% [wt/vol] DOC, 0.1% [wt/vol] SDS), and antibodies (3 μl of rabbit sera and 100 μl of monoclonal antibody tissue culture supernatant) were added. The solutions were incubated at 4°C for at least 3 h, after which 30 μl of Pansorbin (Calbiochem) was added, and the incubation continued for at least 1 h. Immune complexes were then collected by centrifugation, washed three times with wash buffer I (20 mM Tris [pH 7.6], 150 mM NaCl, 5 mM EDTA, 0.1% NP-40) and once with wash buffer II (20 mM Tris [pH 7.6], 0.1% NP-40).
Immunoisolation of virus particles was performed by diluting 450 μl of cleared cell culture supernatant with a similar volume of TNE (20 mM Tris [pH 7.6], 50 mM NaCl, 1 mM EDTA) containing antibodies (3 μl of rabbit serum or 100 μl of monoclonal antibodies). The solution was incubated overnight at 4°C, after which 30 μl of Pansorbin was added to each sample. Incubation was continued for 1 h, after which immune complexes were collected by centrifugation and washed three times with TNE.
For analysis of the proteins, the washed immune complexes were resuspended in 30 μl of Laemmli sample buffer containing 50 mM dithiothreitol (DTT) and analyzed by electrophoresis in SDS-polyacrylamide gels (PAG). The gels were fixed in 10% acetic acid–50% methanol for at least 30 min and incubated for another 30 min in 1 M sodium salicylate. Finally, the gels were dried and subjected to fluorography at −80°C. Radioactivity in protein bands in PAG was quantitated by using a PhosphorImager and ImageQuant (Molecular Dynamics) analysis.
Proteinase K treatment of MHV virions.
After labeling of MHV-A59-infected OST7-1 cells, the culture medium was collected, cleared, and adjusted to 0.01 M Tris (pH 7.8) and 1 mM CaCl2 (proteinase K reaction buffer) by using a 100× concentrated stock solution. Proteinase K was then added from a stock solution to some samples to a concentration of 150 μg/ml, and all samples were incubated for 1 h on ice. The enzymatic treatment was stopped by adding phenylmethylsulfonyl fluoride (PMSF) to a concentration of 7.5 mM while keeping the samples on ice. A one-fifth volume of concentrated lysis buffer was then added to each sample, and immunoprecipitations were performed as detailed above but in the presence of 5 mM PMSF in all solutions.
In vitro transcription and translation.
In vitro transcription reactions were carried out by using T7 RNA polymerase (Boehringer Mannheim) according to the manufacturer's instructions in 50-μl volumes containing 4 μg of PstI-linearized pT7Ts-E. After a 1-h incubation at 37°C, the RNA was purified by chromatography in a Sephadex G50 column and precipitated with ethanol, and the dried pellet was dissolved in 50 μl of TNE.
Translations of the mRNAs (0.8 μl of RNA transcript in a 10-μl reaction) were done for 1 h at 30°C in the Promega rabbit reticulocyte lysate system in the presence or absence of canine microsomal membranes (Promega). The samples were then split and diluted to 1 ml with IP buffer. Antisera (3 μl of the rabbit preE serum or the anti-E serum) were added, and the normal immunoprecipitation procedure was followed.
Immunofluorescence analysis of protein membrane topology.
BHK-21 cells expressing the MHV-A59 E or M protein were surface permeabilized by using Streptolysin O (SLO; purchased from S. Bhakdi, Johannes Gutenberg-Universität, Mainz, Germany). At 7 h postinfection, culture media were taken off and cells were rinsed with SLO buffer (25 mM HEPES [pH 7.4], 115 mM potassium acetate, 2.5 mM Mg2Cl). They were then incubated for 15 min on ice with SLO buffer containing 1 mM DTT and 1 μg of SLO per ml and subsequently washed twice with SLO buffer containing 1 mM DTT. To activate the SLO, the cells were incubated for 30 min at 37°C with prewarmed SLO buffer containing 1 mM DTT. The cells were put on ice and directly rinsed with SLO wash buffer (50 mM HEPES [pH 7.4], 5 mM Mg2Cl, 2 mM EGTA, 50 mM KCl). This washing step was repeated once, and the cells were then washed with PBS and subjected to fixation with 3% paraformaldehyde (PFA) for 30 min. Proteinase K treatment of SLO-permeabilized cells was carried out by preincubation of the cells after the PBS washing step for 5 min with proteinase K buffer, followed by incubation for 30 min on ice with proteinase K buffer containing 50 μg of proteinase K per ml. Cells were then rinsed twice with proteinase K wash buffer (2 mM EGTA, 5 mM EDTA, 1.15 mM PMSF [pH 7.4]) and once with PBS containing 50 mM glycine and 1.15 mM PMSF, after which they were fixed with 3% PFA.
For classical indirect immunofluorescence, permeabilization of all cellular membranes was performed after fixation with 3% PFA by treatment for 5 min with 1% Triton X-100–5% FCS in PBS–50 mM glycine. The cells were washed three times with PBS containing 50 mM glycine and 5% FCS.
Fluorescence labeling of the cells with antibodies was done by incubating them with primary antibodies, followed by fluorescein isothiocyanate (FITC)- or tetramethyl rhodamine isocyanate (TRITC)-conjugated secondary antibodies. The primary antibodies used were rabbit anti-E (1:400), mouse anti-MN (undiluted), and rabbit anti-Mc (1:300) sera. The secondary antibodies used were FITC- or TRITC-conjugated goat anti-mouse and goat anti-rabbit immunoglobulin G (Cappel; 1:100).
Electron microscopy.
Mouse L cells infected with MHV-A59 and BHK-21 cells expressing the E protein were fixed at 5.30 and 4 h postinfection (hpi), respectively, and prepared for cryosectioning as described earlier (21). Thawed cryosections were double labeled with the E antibody at a dilution of 1:100 and with Rab-1 antibodies (36) diluted 1:40. The double labeling was carried out according to the sequential protocol of Slot et al. (39). Embedding of cells in Epon was done as described elsewhere (13).
RESULTS
Kinetics of appearance of the E protein in MHV-A59-infected cells.
Sera raised against purified MHV-A59 are very poor in detecting the E protein. Thus, a fusion protein was prepared in order to obtain protein E-specific antibodies for immunological studies. It consisted of the 34-amino-acid carboxy-terminal domain of MHV-A59 E protein preceded by the amino-terminal 161-residue ectodomain of the EAV GS protein (Fig. 1B). By choosing GS as a fusion partner to enhance immune responses, we aimed to also acquire a polyclonal serum to this EAV protein. For purification purposes, the fusion protein was extended amino terminally with a His-tag. The protein was produced in E. coli and injected into rabbits. The immune serum obtained was found to immunoprecipitate a protein with an apparent molecular mass of 9.8 kDa from lysates of radiolabeled cells infected with MHV-A59 or expressing the viral E gene by using the vaccinia virus T7 system (data not shown). This molecular mass closely corresponds to the predicted size of the E protein (9.6 kDa).
We used the antiserum to evaluate the synthesis of the E protein during MHV infection, comparing it with that of the other viral structural proteins. Cultures of infected cells were therefore labeled for successive 1-h periods with 35S-labeled amino acids. Immunoprecipitates were prepared from the cell lysates by using a polyclonal rabbit serum against MHV-A59 that recognizes predominantly the N, M, and S proteins and by using the rabbit serum against the E protein. Precipitates were analyzed in 15 and 20% PAG, respectively, as shown in Fig. 2A and B. For each of the viral proteins the time course of synthesis was determined by quantitation of the radioactivity in the respective gel segments, the results of which, normalized to allow comparison, are compiled in Fig. 2C. It is clear that all four proteins were synthesized with very similar kinetics. They were all first detected at between 3 and 4 hpi, after which their production rapidly and simultaneously increased, reaching a maximal rate around 6 hpi, which was maintained for several hours. Since the proteins were probably all immunoprecipitated with different efficiencies, these results do not allow an estimation of the relative molar ratios of their synthesis.
Stability of the E protein during infection.
To further study the fate of the E protein in infection, we performed a pulse-chase analysis, during which we again monitored the radioactivity in the E protein in comparison with that in the other structural proteins. In order to obtain an overall picture, we included in our analysis the fraction of the proteins that is released from the cells with virions. For this purpose we prepared total lysates in which cells and culture fluids were combined. Immunoprecipitates of M, S, and N proteins were prepared with the anti-MHV serum and analyzed in a 15% gel (Fig. 3A); E protein precipitated with the E-specific serum was electrophoresed in a 20% gel (Fig. 3B). Quantitations of the radioactivity in the different proteins are graphically represented in Fig. 3C. The data show that the structural proteins synthesized at around 6 hpi turned over with quite similar kinetics. It was deduced from this and other experiments that the proteins had half-lives of approximately 4 h. A similar pattern of turnover was obtained when an otherwise identical experiment was performed in which protein synthesis was blocked after the pulse-labeling by including cycloheximide (0.5 mM) in all chase media (data not shown). This result ruled out the possibility that the decline in the amount of radioactive proteins was caused by a gradual shortage of antibodies during the immunoprecipitation as a result of the continued synthesis of (unlabeled) viral proteins.
Inspection of Fig. 3A reveals the appearance during the chase of a polypeptide (marked by an asterisk) running slightly faster in the gel than the normal set of M proteins. This polypeptide, which could also be observed in the experiment depicted in Fig. 2, appeared concomitantly with the conversion of the unglycosylated M protein (M0) synthesized during the pulse into the different, slower-migrating O-glycosylated forms. Using domain-specific antibodies, we have demonstrated that the polypeptide represents an M protein that lacks its normal amino terminus (data not shown). Its discrete size, which is about 2.4 kDa smaller than that of the M0 form, indicates that it has completely lost the domain that is exposed at the luminal side of intracellular membranes, i.e., at the outside of virions.
Stability of individually expressed E protein.
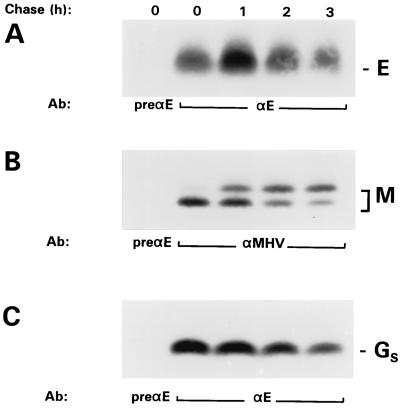
We also studied the stability of the independently expressed E protein in a pulse-chase experiment. Because no E protein appeared to be released into the culture medium (data not shown) only the cell-bound protein was analyzed. For comparison, we similarly expressed in parallel the M protein (known to be stable [34rsqb;) and the EAV GS protein, which we found earlier to be subject to degradation both in EAV-infected BHK-21 cells and when expressed individually in these cells (7). As the results compiled in Fig. 4 show, the M and GS proteins behaved as expected, although the latter protein, immunoprecipitated by the antiserum directed against GS-E fusion protein, appeared to be less prone to turnover in the OST7-1 cells than in the BHK-21 cells used before. The stability of the E protein was comparable with that of the GS protein under these conditions; its level remained constant initially but started to decrease thereafter.
FIG. 4.
Stability of the expressed E protein. vTF7-3-infected OST7-1 cells transfected with pT7Ts-E, pTUM-M, or pAVI02 plasmids containing the E, the M, and the EAV GS genes, respectively, were labeled with 35S-labeled amino acids (80 μCi/10-cm2 dish) for 1 h starting at 6 hpi. Cell lysates were prepared immediately after the labeling or after various chase times as indicated, and immunoprecipitations were carried out with the antisera anti-E (αE), pre-anti-E (preαE), and anti-MHV (αMHV). Proteins were analyzed in 20% PAG.
Membrane integration of E occurs without signal sequence cleavage.
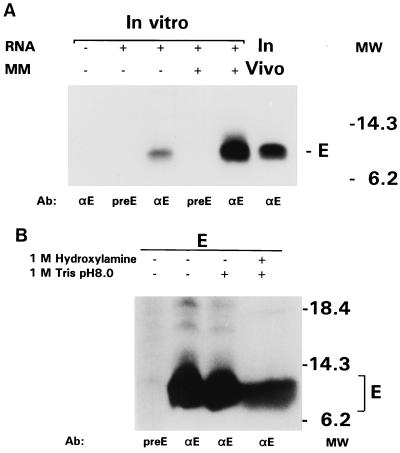
The MHV-A59 E protein contains a rather long hydrophobic domain at its amino terminus which is likely to mediate its membrane integration. To find out whether its membrane insertion involves the functioning of a classical, cleavable signal sequence, we compared the size of the primary translation product with that of the membrane-integrated form. Capped RNA was transcribed in vitro from an E gene plasmid construct and translated in a reticulocyte lysate system in the absence and in the presence of rough microsomal membranes. The labeled products were immunoprecipitated by using the E antiserum, and their electrophoretic mobilities were analyzed in parallel with that of E protein synthesized in transfected cells by using the vaccinia virus expression system. Figure 5A shows that the E proteins synthesized in vitro and in cells comigrated. Apparently, no signal sequence is cleaved during or after membrane integration of the protein unless the size reduction that would accompany such cleavage is compensated for by a secondary modification, for which we have no indications. The MHV-A59 E protein sequence does not contain an N-glycosylation motif. Moreover, as we show in Fig. 5B, we were unable to confirm the conclusion made by Yu et al. (49), on the basis of hydroxylamine treatment, that the E protein is acylated: in our experiment the protein remained unaffected. Attempts to label the protein in infected cells with 3H-labeled palmitic acid were equally unsuccessful (not shown). These observations are consistent with those of others who were similarly unable to label the TGEV E protein in infected cells with 3H-labeled palmitic acid (12).
FIG. 5.
E protein membrane integration without cleavage of a signal sequence. (A) RNA transcribed in vitro from plasmid pT7Ts-E was translated in a rabbit reticulocyte lysate in the presence or absence of canine microsomal membranes (MM). In parallel, vTF7-3-infected OST7-1 cells transfected with pT7Ts-E were labeled (80 μCi/10-cm2 dish) from 6 to 7 hpi and then lysed. Immunoprecipitations were carried out with the anti-E (αE) serum and the preE serum, and the proteins were analyzed in 20% PAG. (B) A similar OST7-1-derived immunoprecipitate of the E protein was split up into two parts, which were both incubated in 1 M Tris-Cl (pH 8.0), one of which also contained 1 M hydroxylamine. Samples were then diluted with Laemmli sample buffer and analyzed in 20% PAG. Untreated control immunoprecipitates prepared with anti-E and preE serum were included in the analysis for comparison. Note that the presence of hydroxylamine caused a broadening of the protein band in the right lane.
Topology of the membrane-assembled E protein.
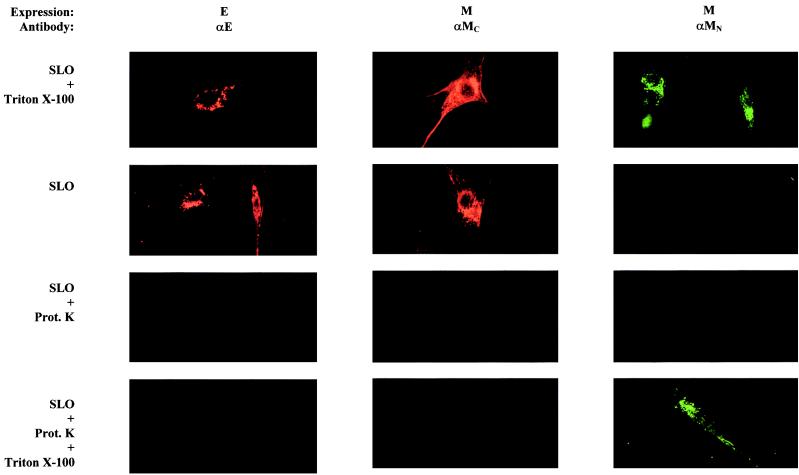
Besides the lack of a cleaved signal sequence at its amino terminus the E protein also lacks the typical anchor sequence occurring in the carboxy-terminal region of so many other membrane proteins. Rather, its hydropathy plot suggests that the polypeptide is buried within the lipid bilayer for most of its amino-terminal 60 residues. To determine the disposition of the residual more-hydrophilic carboxy terminus, we took an immunofluorescence approach based on the availability of an antiserum specifically recognizing this part of the protein. In this assay cells were permeabilized selectively in their plasma membrane by using SLO to allow intracellular access of antibodies. The assay was first tested with cells expressing the M protein whose membrane topology has been well established (33, 35): its amino- and carboxy-terminal domains are exposed luminally and cytoplasmically, respectively, and antibodies directed to these domains are available. Accordingly, surface-permeabilized cells could not be stained with the anti-MN antibodies unless the intracellular membranes were also permeabilized by using Triton X-100 (Fig. 6). In contrast, the anti-MC antibodies readily stained the surface-permeabilized cells, thus confirming the cytoplasmic exposure of the carboxy terminus. Consistently, staining was fully abrogated by prior treatment of the permeabilized cells with proteinase K, a treatment that only affected the cytoplasmically exposed protein domains since the amino terminus of the M protein remained intact and could still be visualized by the anti-MN antibodies after additional Triton X-100 permeabilization. This latter treatment did not enable staining with the anti-MC antibodies, indicating that the M protein assumes one defined membrane topology, in contrast to the TGEV M protein, which was reported to take two alternative orientations (31).
FIG. 6.
Location of the carboxy terminus of the membrane-integrated E protein. BHK-21 cells expressing the M protein or the E protein were permeabilized by using either SLO or Triton X-100 and stained for immunofluorescence with the indicated antibodies. In some cases, the SLO-permeabilized cells were first treated with proteinase K (Prot.K) before the staining, while in other cases the SLO-permeabilized and proteinase K-treated cells were additionally permeabilized by using Triton X-100 and then incubated with the antibodies. αMC, peptide antiserum to the M protein's C-terminal tail; αMN, monoclonal antibody J1.3 specific for the M protein's N terminus.
When the immunofluorescence assay was applied to cells expressing the E protein, it became clear that its carboxy terminus protrudes into the cytoplasm. The domain was accessible to the anti-E antibodies after surface permeabilization (Fig. 6). Proteinase K treatment completely abolished the staining, and subsequent permeabilization with Triton X-100 did not reveal any “hidden” carboxy-terminal domains, a result consistent with a unique membrane topology.
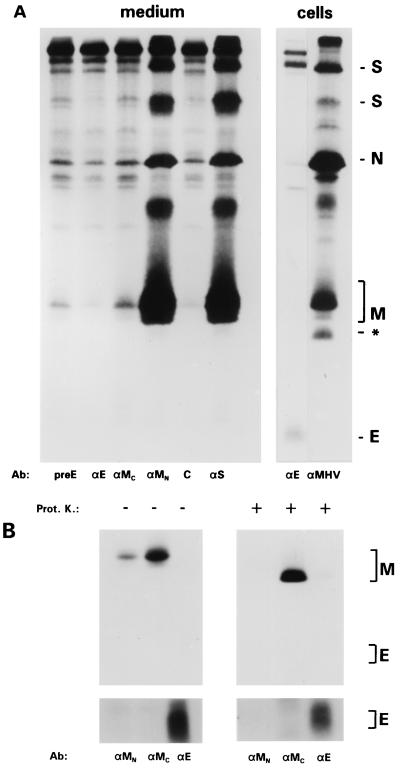
A cytoplasmic exposure of the E protein's carboxy terminus would topologically correlate with a disposition of this domain on the inside of MHV particles. Accordingly, we were unable to label VLPs, obtained by expressing the MHV M and E proteins, in an immunogold labeling with the anti-E antibodies, as judged under the electron microscope (data not shown). To further corroborate this point, we also used a biochemical approach. Because of its great sensitivity, we performed an immunoisolation of radioactively labeled virus particles from the culture medium of MHV-A59-infected cells by using various antibodies. As the protein pattern of Fig. 7A shows, the S-specific antibodies (Fig. 7A, αS) not only isolated S protein but in addition all other structural proteins M, N, and E, the latter one being visible only after prolonged exposure (not shown). Similarly, viral particles could be isolated with the anti-MN antibodies but not with the anti-MC antibodies, a finding consistent with the known exterior and interior disposition of the corresponding M domains, respectively. As expected, no trace of viral particles was detected when the anti-E serum was used in the immunoisolation, confirming the interior disposition of the E protein's carboxy terminus in virions.
FIG. 7.
Topology of the E protein in MHV-A59 virions: the carboxy-terminal domain is not exposed on the outside. (A) Immunoaffinity isolation of virions. OST7-1 cells grown in a 10-cm2 dish and infected with MHV-A59 were labeled with 35S-labeled amino acids at 6 to 9 hpi, after which the cells were lysed and processed for immunoprecipitation with anti-MHV (αMHV) and anti-E (αE) (right panel). The culture medium containing released virus particles was clarified, and aliquots were first incubated with the antibodies as indicated (for explanation, see previous figures; αS, monoclonal antibody J7.6 recognizing an epitope in the S ectodomain; C, control monoclonal antibody) and subsequently with Staphylococcus aureus bacteria to adsorb the antibodies and associated viral particles, which were analyzed in 20% PAG. (B) Protease protection analysis. The culture medium of a similarly labeled 10-cm2 dish of infected cells was split and either treated with proteinase K (Prot. K) or incubated without enzyme. Concentrated lysis buffer was then added, and standard immunoprecipitations were carried out with the antibodies indicated (for explanations, see the legends to previous figures). Proteins were analyzed in 20% PAG. From the upper panel, which was exposed for 4 h, the region containing the E protein is shown after a longer exposure (4 days) in the lower panel.
Due to its very low abundance, the E protein is difficult to detected in MHV-A59 particles. We nevertheless analyzed the effect of proteolytically removing the exposed domains of the viral membrane proteins. Figure 7B shows that treatment of 35S-labeled virions eliminated the epitope present in the amino terminus of the M protein recognized by the monoclonal J1.3 (αMN). It also reduced the size of the polypeptide by about 2.5 kDa as shown previously (33). The E protein seemed unaffected. Its appearance as a diffuse band, visible only after long exposure of the gel, did not change as a result of the treatment. The mere fact that the protein remained precipitable by the anti-E antibody implies that the carboxy terminus was indeed protected. In addition, it shows that no other part(s) of the protein is significantly exposed on the outside of virions.
Electron microscopic observations.
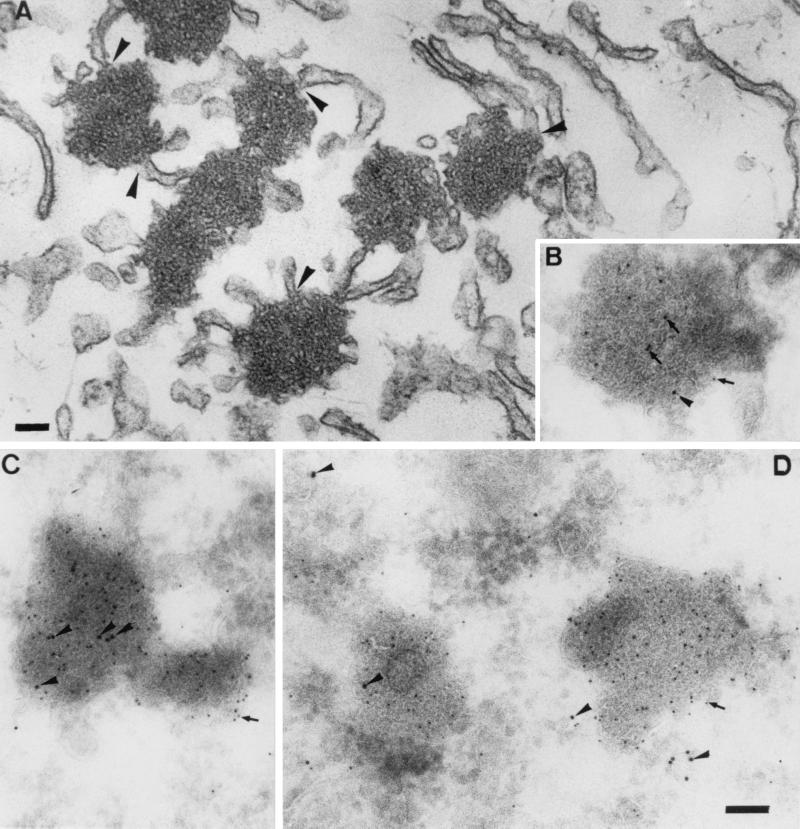
One of the marked observations made during the immunofluorescence studies described above was the peculiar punctate staining pattern of the expressed E protein. To study this aspect in more detail, BHK-21 cells expressing the protein were prepared for and then analyzed by electron microscopy. It appeared that the E protein induced the formation of characteristic electron-dense structures (Fig. 8A) that were not seen in control cells, for instance, when the M protein was expressed similarly. They were, however, also observed in MHV-A59-infected L cells (as shown after cryosectioning in Fig. 8B) and are actually very reminiscent of the tubular structures described in MHV-A59-infected cells many years ago (4). They seem to consist of masses of tubular, smooth membranes with much curvature that form complicated networks. The membrane clusters are relatively homogeneous in that other organelles are excluded. Continuities with the endoplasmic reticulum (ER) are, however, often observed. The structures were obviously induced by the E protein since they labeled heavily with the anti-E antibody (Fig. 8B to D). In fact, in highly expressing cells the E protein often appeared to localize almost exclusively to them. Interestingly, the tubular structures are part of the ER-intermediate compartment (IC) network, as was demonstrated by their colabeling on cryosections with a marker for these compartments, Rab-1 (Fig. 8B to D; see also references 14 and 36).
FIG. 8.
Electron microscopic analysis. (A) Epon section of BHK-21 cell expressing the E protein and fixed at 4 h posttransfection. The E protein induces the formation of electron-dense membrane structures that are often continuous with the rough ER (arrowheads). (B to D) The same structures in thawed cryosections double labeled for E (5-nm gold; arrows) and Rab-1 (10-nm gold; arrowheads). Panels: B, an MHV-infected cell fixed at 5.30 hpi, C and D, BHK-21 cells expressing the E protein. Bars, 100 nm.
DISCUSSION
The remarkable role of the E protein in coronavirus assembly prompted the detailed characterization of the envelope protein described here. E protein emerges from these studies as a unique protein. The unglycosylated polypeptide appears in MHV-A59-infected cells synchronously with all the other viral structural proteins. It is assembled in the ER membrane by virtue of an uncleaved signal sequence assuming a complex membrane structure which leaves its carboxy terminus in the cytoplasm. In virions this domain is oriented inwards, facing the nucleocapsid. When expressed independently, the E protein accumulates in and induces the coalescence of the membranes of the ER-IC, giving rise to characteristic structures that also appear in infected cells.
The E protein appears to integrate into the ER membrane without modifications. Membrane insertion is mediated by a signal within the large hydrophobic domain, most likely by the 28-residue stretch preceding the conserved lysine at position 38. No cleavage of this domain is predicted nor was it observed. Hence, our biochemical analyses of permeabilized cells expressing the protein and of viral particles lead to a picture of a largely membrane-embedded E protein, of which only the ∼23-residue hydrophilic tail is exposed cytoplasmically and on the inside of virions. No part of the protein was detectably exposed on the virion outside, the topological equivalent of the ER lumen. The amino terminus may be oriented to either side of the membrane. Other, more biophysical methods will be required to determine the fine structure of the membrane-integral part of the molecule which is sufficiently long to span the lipid bilayer twice. It will be interesting to find out to which side of the membrane the conserved lysine (position 38) is oriented, what the disposition is of the conserved cysteines immediately downstream of this lysine, and what structural position the proline at position 54 takes that is absolutely conserved among all coronavirus E proteins (10, 38).
In earlier immunofluorescence studies the E protein has been detected at the surface of infected cells. With antibodies prepared against the whole E protein, positive but weak staining of unfixed cells infected with infectious bronchitis virus was observed (40). Surface staining was also reported for MHV- (49) and TGEV-infected cells, as well as for insect cells expressing the TGEV E protein by a baculovirus vector (12). The antibodies used in these studies were directed against the protein's carboxy terminus, leading Godet et al. (12) to the suggestion that this region of the molecule is translocated across the membrane, a finding which is at variance with our observations. The reasons for this discrepancy are unknown and remain to be elucidated.
In MHV-infected cells the structural proteins are synthesized from subgenomic mRNAs. These mRNAs are synthesized in different molar amounts but at constant relative ratios (17, 24). Except for the smallest mRNA, encoding just the N protein, they are all structurally polycistronic but express only their 5′-terminal open reading frame (ORF). The E protein, however, is translated from a downstream ORF by internal ribosome entry (3, 44). Our quantitations of the synthesis of the S, N, M, and E proteins during infection show similar kinetics for all of them. Although the proteins are probably produced in very different molar amounts, their time courses of synthesis were indistinguishable.
In one of the first extensive electron microscopic studies of coronavirus-infected cells David-Ferreira and Manaker (4) described the appearance of structures “formed by closely interwoven, membrane-limited tubules.” These structures, which were termed tubular bodies, were induced by the MHV-A59 infection. They appear to be very similar to what we found in our present studies, not only in infected cells but also in response to the independent expression of the E protein. This observation thus links the induction of the structures directly to the E protein. Moreover, their colabeling with antibodies to E and to the ER-IC marker Rab-1 suggests that they are derived by the coalescence of these pre-Golgi membranes. Although its precise cellular localization was beyond the scope of this study, our preliminary observations suggest that the expressed E protein accumulates in membranes of the ER-IC. We and others have shown earlier that budding of coronaviruses occurs at these particular membranes (18, 21, 45, 46). The reasons for this have so far remained obscure because neither of the major viral envelope proteins was found to localize to these membranes (20, 29). If the E protein by itself indeed localizes to pre-Golgi membranes, this would most likely provide the explanation. By its interactions with the M protein the E protein might retain M protein molecules in the early compartments, allowing the accumulation of the large lateral assemblies where viral particles are formed (22, 29).
While numerous viruses have been shown to variously induce novel structures in infected cells, rarely have these structures been assigned to the action of a particular viral protein. An interesting exception are the tubular networks of smooth membranes that were reported by Hobman et al. (15, 16) to arise in cells upon expression of the rubella virus E1 glycoprotein. The characteristics of these structures are very similar to the ones described here, notably with respect to their morphology, their continuity with ER membranes, their labeling with antibodies to ER-IC marker proteins, and their insensitivity to brefeldin A (data not shown). The E1-induced tubular networks were purified from cells by immunoisolation and shown to likely correspond to hypertrophied ER exit sites (16).
The convoluted appearance of the tubular membranes that we observed in response to the E protein may point to an important biological feature of the protein in coronavirus morphogenesis. It suggests that the E protein has a tendency to induce curvature into membranes. Earlier we hypothesized that this might be one of two possible roles that the protein might have during the budding of coronaviral particles (48). The alternative role was in the closing of the neck of the nascent particle, causing the pinching off of the virion. It is still too early to decide whether the protein indeed serves one of these functions. Recent support for an important function of the E protein in virion morphogenesis comes from analyses of MHV-A59 E gene mutants (10). Mutations introduced into the E protein's hydrophilic tail by targeted RNA recombination yielded viruses that were markedly thermolabile, suggesting that there was a flaw in their structure. The particles of one of the viruses were viewed by electron microscopy and appeared to be aberrant and heterogeneous in their morphology. Instead of the normal rounded structures, most virions had elongated, tubular forms, often pinched at multiple points, producing dumbbell-shaped structures. The pictures suggest that the E protein indeed is important for creating the membrane curvature needed to acquire the rounded, stable, and infectious particle phenotype.
The occurrence of small membrane proteins appears to be quite a general feature of enveloped RNA viruses. For some of these a function has been established such as for the ion-channel M2 protein of influenza virus (30) and for the alphavirus 6K protein which is important in the final assembly and budding of virions (25, 27). Interestingly, we recently discovered that a small membrane protein also occurs in arteriviruses (41), which constitute another genus in the order of Nidovirales (8). This 67-residue E protein is a structural component of EAV. The polypeptide is very hydrophobic, with a basic carboxy terminus, and is membrane associated in infected cells. Importantly, the protein appeared to be essential for the production of infectious viral particles. In view of these findings it is surprising that no small membrane protein has so far been identified in toroviruses, the second family of viruses belonging to the Coronaviridae. While these viruses do have an S protein and an M protein very similar to those of coronaviruses, no E protein homologue seems to be encoded. This suggests that the budding of these viruses occurs differently from coronaviruses or that the function of the E protein is expressed otherwise. We expect that our continued comparative studies of these different nidoviruses will eventually provide more detailed insight into the various ways these viruses assemble their particles.
ACKNOWLEDGMENT
This work was financially supported in part by Intervet International B.V., which we gratefully acknowledge.
REFERENCES
- 1.Abraham S, Kienzle T E, Lapps W E, Brian D A. Sequence and expression analysis of potential nonstructural proteins of 4.9, 4.8, 12.7, and 9.5 kDa encoded between the spike and membrane protein genes of the bovine coronavirus. Virology. 1990;177:488–495. doi: 10.1016/0042-6822(90)90513-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredenbeek P. Nucleic acid domains and proteins involved in the replication of coronaviruses. Ph.D. thesis. Utrecht, The Netherlands: Utrecht University; 1990. [Google Scholar]
- 3.Budzilowicz C J, Weiss S R. In vitro synthesis of two polypeptides from a nonstructural gene of coronavirus mouse hepatitis virus strain A59. Virology. 1987;159:509–515. doi: 10.1016/0042-6822(87)90293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David-Ferreira J F, Manaker R A. An electron microscope study of the development of a mouse hepatitis virus in tissue culture cells. J Cell Biol. 1965;24:57–78. doi: 10.1083/jcb.24.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan C A M, Roestenberg P, de Wit M, de Vries A A F, Nilsson T, Vennema H, Rottier P J M. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J Biol Chem. 1998;273:29905–29914. doi: 10.1074/jbc.273.45.29905. [DOI] [PubMed] [Google Scholar]
- 6.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries A A F, Raamsman M J B, van Dijk H A, Horzinek M C, Rottier P J M. The small envelope glycoprotein (Gs) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J Virol. 1995;69:3441–3448. doi: 10.1128/jvi.69.6.3441-3448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesize bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godet M, Haridon R L, Vautherot J-F, Laude H. TGEV coronavirus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths G. Fine structure immunocytochemistry. Heidelberg, Germany: Springer-Verlag; 1993. [Google Scholar]
- 14.Griffiths G, Ericsson M, Krijnse Locker J, Nilsson T, Soeling H D, Tang B L, Wong S H, Hong W. Ultrastructural localization of the mammalian KDEL receptor in cultured cells and tissues. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobman T C, Woodward L, Farquhar M G. The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J Cell Biol. 1992;18:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobman T C, Zhao B, Chan H, Farquhar M G. Immunoisolation and characterization of a subdomain of the endoplasmic reticulum that concentrates proteins involved in COPII vesicle biogenesis. Mol Biol Cell. 1998;9:1265–1278. doi: 10.1091/mbc.9.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs L, Spaan W J M, Horzinek M C, van der Zeijst B A M. Synthesis of subgenomic mRNAs of mouse hepatitis virus is initiated independently: evidence from UV transcription mapping. J Virol. 1981;39:401–412. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krijnse Locker J, Rose J K, Horzinek M C, Rottier P J M. Membrane assembly of the triple-spanning coronavirus M protein: individual transmembrane domains show preferred orientation. J Biol Chem. 1992;267:21911–21918. doi: 10.1016/S0021-9258(19)36699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krijnse Locker J, Griffiths G, Horzinek M C, Rottier P J M. O-Glycosylation of the coronavirus M protein: differential localization of sialyltransferases in N- and O-glycosylation. J Biol Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krijnse Locker J, Ericsson M, Rottier P J M, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krijnse Locker J, Opstelten D-JE, Ericsson M, Horzinek M C, Rottier P J M. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J Biol Chem. 1995;270:8815–8821. doi: 10.1074/jbc.270.15.8815. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Leibowitz J L, Wilhelmsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D X, Inglis S C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loewy A, Smyth J, von Bonsdorff C-H, Liljeström P, Schlesinger M J. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol. 1995;69:469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opstelten D-JE, de Groote P, Horzinek M C, Vennema H, Rottier P J M. Disulfide bonds in folding and transport of the mouse hepatitis virus glycoproteins, J. Virol. 1993;67:7394–7401. doi: 10.1128/jvi.67.12.7394-7401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opstelten D-JE, Raamsman M J B, Wolfs K, Horzinek M C, Rottier P J M. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 31.Risco C, Anton I M, Sune C, Pedregosa A M, Martin-Alonso J M, Parra F, Carrascosa J L, Enjuanes L. Membrane protein molecules of transmissible gastroenteritis virus also expose the carboxy-terminal region on the external surface of the virion. J Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottier P J M, Spaan W J M, Horzinek M C, van der Zeijst B A M. Translation of three mouse hepatitis virus strain A59 subgenomic RNAs in Xenopus laevis oocytes. J Virol. 1981;38:20–26. doi: 10.1128/jvi.38.1.20-26.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rottier P J M, Brandenburg D, Armstrong J, van der Zeijst B A M, Warren G. Assembly in vitro of a spanning membrane protein of the endoplasmic reticulum: the E1 glycoprotein of coronavirus MHV-A59. Proc Natl Acad Sci USA. 1984;81:1421–1425. doi: 10.1073/pnas.81.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rottier P J M. The coronavirus membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 35.Rottier P J M, Welling G W, Welling-Wester S, Niesters H G M, Lenstra J A, van der Zeijst B A M. Predicted membrane topology of the coronavirus protein E1. Biochemistry. 1986;25:1335–1339. doi: 10.1021/bi00354a022. [DOI] [PubMed] [Google Scholar]
- 36.Saraste J, Lahtinen U, Goud B. Localization of the small GTP-binding protein rab1p to early compartments of the secondary pathway. J Cell Sci. 1995;108:1541–1552. doi: 10.1242/jcs.108.4.1541. [DOI] [PubMed] [Google Scholar]
- 37.Siddell S G. The Coronaviridae. New York, N.Y: Plenum Press; 1995. [Google Scholar]
- 38.Siddell S G. The small-membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 181–189. [Google Scholar]
- 39.Slot J W, Geuze H G, Gigengack S, Lienhard G E, James D E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith A R, Boursnell M E G, Binns M M, Brown T D K, Inglis S C. Identification of a new membrane-associated polypeptide specified by the coronavirus infectious bronchitis virus. J Gen Virol. 1990;71:3–11. doi: 10.1099/0022-1317-71-1-3. [DOI] [PubMed] [Google Scholar]
- 41.Snijder E J, van Tol H, Pedersen K W, Raamsman M J B, de Vries A A F. Identification of a novel structural protein of arteriviruses. J Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spaan W J M, Rottier P J M, Horzinek M C, van der Zeijst B A M. Isolation and identification of virus specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59) Virology. 1981;108:424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutter G, Ohlman M, Erfle V. Non-replicating vaccinia virus vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 44.Thiel V, Siddell S G. Internal ribosome entry in the coding region of murine hepatitis virus mRNA5. J Gen Virol. 1994;75:3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]
- 45.Tooze J, Tooze S A, Warren G. Replication of coronavirus MHV-A59 in Sac− cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 46.Tooze S A, Tooze J, Warren G. Site of addition of N-acetylgalactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J Cell Biol. 1988;106:1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tung F Y T, Abraham S, Sethna M, Hung S L, Sethna P, Hogue B G, Brian D A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-JE, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope proteins. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]