Abstract
Expression studies have shown that the coronavirus small envelope protein E and the much more abundant membrane glycoprotein M are both necessary and sufficient for the assembly of virus-like particles in cells. As a step toward understanding the function of the mouse hepatitis virus (MHV) E protein, we carried out clustered charged-to-alanine mutagenesis on the E gene and incorporated the resulting mutations into the MHV genome by targeted recombination. Of the four possible clustered charged-to-alanine E gene mutants, one was apparently lethal and one had a wild-type phenotype. The two other mutants were partially temperature sensitive, forming small plaques at the nonpermissive temperature. Revertant analyses of these two mutants demonstrated that the created mutations were responsible for the temperature-sensitive phenotype of each and provided support for possible interactions among E protein monomers. Both temperature-sensitive mutants were also found to be markedly thermolabile when grown at the permissive temperature, suggesting that there was a flaw in their assembly. Most significantly, when virions of one of the mutants were examined by electron microscopy, they were found to have strikingly aberrant morphology in comparison to the wild type: most mutant virions had pinched and elongated shapes that were rarely seen among wild-type virions. These results demonstrate an important, probably essential, role for the E protein in coronavirus morphogenesis.
Coronaviruses are enveloped positive-strand RNA viruses that contain, at a minimum, four structural proteins (39). One of these, the nucleocapsid protein (N), resides in the interior of the virion, encapsidating the large (27- to 32-kb) viral genome. Surrounding the nucleocapsid is an envelope derived from the membrane of the compartment between the endoplasmic reticulum and the Golgi, into which virion budding takes place. Embedded in this envelope are the other three structural proteins: the spike glycoprotein (S), the membrane glycoprotein (M), and the small membrane or envelope protein (E; formerly m or sM). Two additional structural proteins, a hemagglutinin-esterase glycoprotein (HE) (7) and the product of the internal open reading frame (ORF) of the N gene (I protein) (13, 38), are absent in a number of coronavirus species or strains and are not essential for infectivity.
For the membrane-bound proteins, the importance of S in receptor binding and fusion and of M as the predominant constituent of virion architecture has been understood for some time. The significance of E, however, has come to be appreciated only relatively recently. E originally had the status of a hypothetical product of a small ORF revealed by cDNA sequencing (5, 40). Subsequently, it was shown to be translated both in vitro (8, 25) and in vivo (1, 16, 23, 25). This latter point, that E protein is actually expressed during infection, could not be presumed a priori because in mouse hepatitis virus (MHV) and avian infectious bronchitis virus (IBV), the coding region for E is found as the most downstream ORF in a bi- or tricistronic mRNA (5, 40). Next, contrary to most earlier assumptions, it was shown for IBV (26), for porcine transmissible gastroenteritis virus (16), and finally for MHV (47) that the E protein is a coronavirus structural protein. This finding had probably eluded prior analyses because of the small size (9 to 12 kDa) and low abundance of E protein in virions.
The potential role of E protein in viral infection remained speculative until landmark studies by Vennema et al. (42) and Bos et al. (4) examined the assembly of virus-like particles resulting from the intracellular expression of MHV proteins in various combinations. The surprising outcome of these investigations was that the E and M proteins were shown to be both necessary and sufficient for the formation and extracellular release of particles appearing structurally identical to MHV virions (except for the absence of S protein spike projections on their surface if this protein was not coexpressed). This discovery established a new paradigm for enveloped-virus assembly and motivated us to seek genetic evidence supporting such a critical role for E protein in the whole virus.
The genetic manipulation of MHV and other coronaviruses currently is less straightforward than for other positive-strand RNA viruses. For the large genomes of members of this family, it has not yet been possible to construct full-length cDNA clones from which, in principle, infectious RNAs could be transcribed. We have been able to generate site-directed mutations in MHV by targeted recombination between transfected synthetic donor RNA species and recipient mutant viruses that can be selected against (13, 14, 19, 22, 32, 33). In the present study, this technique was taken a step further in that engineered mutants were identified by screening rather than by selection, thereby allowing the construction of mutants that were almost as defective as the parental virus from which they were derived. We chose to create mutants of the E protein of MHV by clustered charged-to-alanine mutagenesis, a genetic strategy that comprehensively surveys the effects of localized surface alterations on a given protein (2, 3, 44). It is based on the supposition that clusters of charged amino acid residues are more likely to appear on the surface, rather than the interior, of proteins and that these often make strong contributions to protein-protein interactions. Such interactions are expected to be destabilized by the replacement of each member of a charge cluster with the small aliphatic side chain of alanine, a substitution which ought to have benign consequences with respect to normal protein folding. Clustered charged-to-alanine mutagenesis has yielded a remarkably high proportion of conditional-lethal mutants (including temperature-sensitive lethal mutants) in studies of actin (44), β-tubulin (34), the poliovirus polymerase (11) and the vaccinia virus G2R protein (17); however, some proteins may be intractable to this approach (45). We constructed MHV E protein mutants by applying a fixed algorithm in which, for each instance where two or more charged amino acids appeared within a sliding window of 5 residues, a mutant was constructed that replaced all of these charged residues with alanine (44). Two of the mutants obtained were partially temperature sensitive and were dramatically defective in virion assembly.
MATERIALS AND METHODS
Virus and cells.
Wild-type, mutant, and revertant virus stocks of all MHV strains were propagated in mouse 17 clone 1 (17Cl1) cells. The sources of strains MHV-1 and MHV-3 were described previously (30). It should be noted that our laboratory strain of MHV-3 differs from the Bicêtre strain of MHV-3 (9). MHV-2 and MHV-DVIM were generously provided by Ehud Lavi (University of Pennsylvania School of Medicine) and Kathryn Holmes (University of Colorado Health Sciences Center), respectively. Plaque titer determinations and plaque purifications were carried out with mouse L2 cells, which were also maintained in spinner culture for RNA transfection via electroporation, as described previously (29).
Cloning and sequencing of the E gene of various MHV strains.
A region encompassing the E genes of MHV-1, MHV-2, MHV-3, and MHV-DVIM RNA isolated either from infected cells or from purified virus was amplified by reverse transcription followed by PCR (RT-PCR), with primers corresponding to nucleotides (nt) 147 to 164 of gene 5a and nt 244 to 261 of the M gene. Although these primers were based on the known sequence of MHV-A59, a 710-bp PCR product was successfully obtained for each of the four other strains and was cloned into a pCR TA cloning vector (Invitrogen). A consensus nucleotide sequence was determined from three or four clones for each MHV strain.
Plasmid constructs.
T7 transcription vectors generated for the transfer of mutations into the E gene of MHV were derived from pCFS8 (14), which encodes a pseudo-defective interfering (DI) RNA that includes the complete S gene and all of the MHV-A59 genome downstream of S through the poly(A) tail. As detailed previously (14), in this region pCFS8 contains two differences from the authentic MHV sequence: (i) a coding-silent change in the S gene that eliminates a HindIII site, and (ii) a phenotypically silent 19-nt tag at the start of gene 4 that contains an XbaI site.
Clustered charged-to-alanine mutations 1 to 4 were initially created in pCFS1 (14), a precursor of pCFS8 that contains the segment extending from the MluI site at nt 3262 of the S gene through the KpnI site at nt 394 of the M gene (see Fig. 3). Mutations 1 and 2 (see Fig. 2) were each generated by a single step of PCR mutagenesis followed by replacement of the BstEII-EcoRV fragment of pCFS1 with the BstEII blunt-end fragment of the mutant PCR product to yield pCFS9 and pCFS10, respectively. Mutations 3 and 4 were generated by splicing overlap extension PCR mutagenesis (18), and the BstEII-EcoRV fragment of each mutant PCR product was used to replace the corresponding segment of pCFS1 to yield pCFS11 and pCFS15, respectively. Mutations 1 to 4 were then transferred into the pseudo-DI vector by incorporation of the MluI-EagI fragment of pCFS9 or the XbaI-EcoRV fragment of pCFS10, pCFS11, or pCFS15 in place of the corresponding fragment of pCFS8 to produce pFF45-pFF48, respectively.
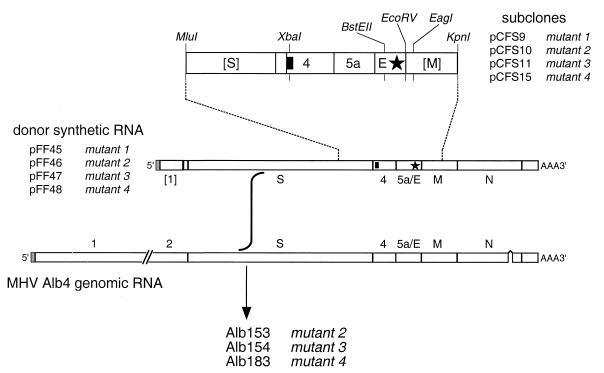
FIG. 3.
Incorporation of clustered charged-to-alanine mutations into the MHV genome by targeted recombination. Sets of mutations engineered into subclones pCFS9, pCFS10, pCFS11, and pCFS15 were transferred to pCFS8 (14) to produce transcription vectors pFF45, pFF46, pFF47, and pFF48, respectively, which encode pseudo-DI RNAs comprising a 5′ segment of the MHV genome fused, at the start of the S gene, to the entire 3′ end of the genome. E gene mutations are represented by a star; the solid rectangle indicates the 19-nt tag at the start of gene 4. Restriction sites shown are those relevant to plasmid construction, as detailed in Materials and Methods. Brackets designate gene fragments rather than entire genes.
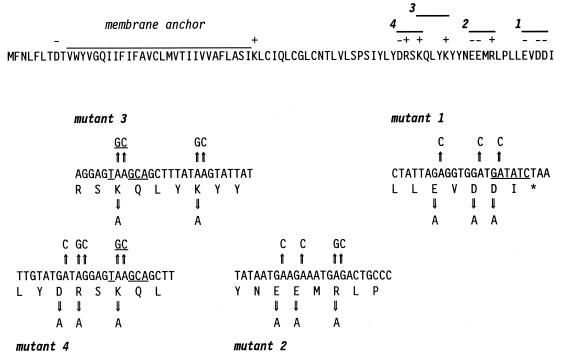
FIG. 2.
Strategy for mutagenesis of the MHV-A59 E gene. The putative membrane anchor, charged amino acid residues, and four possible clustered charged-to-alanine mutants are indicated above the E protein sequence. Beneath are shown the base changes made in transcription vectors to change charged residues to alanines. The EcoRV site near the end of the E coding sequence and the FspI site created by the mutations in mutant 3 and mutant 4 are underlined.
Second-site reverting mutations, in conjunction with the original mutations of mutant 2 (Alb154), were also introduced into MHV via transcription vectors derived from pCFS8. For the construction of these, total RNA isolated from cells infected with Alb154RevA, Alb154RevB, Alb154RevD, or Alb154RevI was reverse transcribed under standard conditions (35) with a random primer, p(dN)6 (Boehringer Mannheim). The 170-bp region from gene 5a through the M gene (see above) of each cDNA was amplified by PCR, restricted with BstEII and EcoRV, and, together with the XbaI-BstEII fragment of pCFS8, incorporated in place of the XbaI-EcoRV fragment of pCFS8 in a three-way ligation to produce plasmids pFF61 to pFF64.
Standard techniques were used for all recombinant DNA manipulations (35). The composition of all constructed plasmids was verified by restriction analysis; all PCR-generated regions and newly created junctions of each plasmid were verified by DNA sequencing by the method of Sanger et al. (36) with modified T7 DNA polymerase (Sequenase; U.S. Biochemical Co.) or by automated sequencing with an Applied Biosystems 373A or 377 DNA sequencer.
Targeted recombination and identification of recombinant mutant viruses.
E gene mutations were transduced into the MHV genome by the targeted recombination method previously used to incorporate mutations into the S (14), M (10), and N (13, 32, 33) genes, gene 4 (14), and the 3′ untranslated region (19, 22). Mutant viruses were obtained as progeny recombinants arising from transfection of synthetic pseudo-DI RNAs from HindIII-truncated pFF45-48 and pFF61-64 into cells infected with the temperature-sensitive and thermolabile N gene mutant, Alb4. Details of RNA synthesis and transfection were as described previously (14, 29).
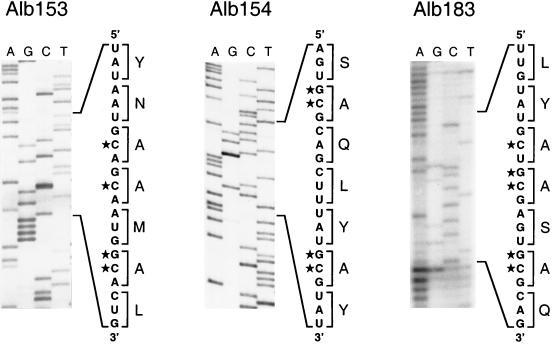
Of the four clustered charged-to-alanine mutants, only mutant 2 was selected as a recombinant capable of forming large (i.e., wild-type-size) plaques at 39°C, the nonpermissive temperature for Alb4. Three candidate plaques for this mutant were analyzed by RT-PCR with a primer pair flanking the locus of the Alb4 deletion and with a primer pair flanking the gene 4 tag (see Fig. 3). The repair of the Alb4 deletion was ascertained by the size of the PCR product from the former (22). The PCR product from the latter was digested with XbaI to determine the presence of the gene 4 tag. Following sequence verification, a recombinant from this set was designated Alb153.
Since targeted recombination with donor RNA containing the other three mutations did not yield large-plaque mutants at 39°C, a screening approach was used for their identification. Following heat treatment to counterselect Alb4 parental virus (22), surviving plaques obtained at 33°C were combined in multiple pools of 10 plaques and RNA was isolated from a cell monolayer infected with each pool and was analyzed by RT-PCR. Recombinants were detected with primer pairs in which one primer originated within the heterologous 19-nt gene 4 tag or in which one primer originated within the region that is deleted in the Alb4 N gene. Plaques from pools giving a positive PCR signal for one of these markers were subsequently analyzed by RT-PCR.
Individual candidates for mutant 3 (20 plaques) and mutant 4 (40 plaques) were then screened for the gene 4 tag or for repair of the Alb4 deletion with primer pairs flanking or originating within these markers, as above. Positive candidates were then screened by RT-PCR amplification of the E gene and tested for the presence of an FspI site created by the mutations in mutant 3 or mutant 4 (see Fig. 2). One mutant of each type was found in this search, and following sequence verification, they were designated Alb154 (mutant 3) and Alb183 (mutant 4). A similar screening procedure was used to search for mutant 1. In this case, the RT-PCR product derived from the E gene was tested for the loss of the EcoRV site that should be disrupted by one of the mutations in this mutant (see Fig. 2). In multiple trials, no recombinant meeting this criterion was found.
Final confirmation of the composition of each of the three successful recombinants was obtained by sequencing purified genomic RNA to establish the presence of the E gene mutations, the gene 4 tag, and the region that is absent in the Alb4 N gene deletion. MHV recombinants were purified by polyethylene glycol precipitation followed by two cycles of equilibrium centrifugation on preformed potassium tartrate-glycerol gradients, and viral genomic RNA was isolated exactly as described previously (13, 22). For the isolation of total cytoplasmic RNA from virus-infected 17Cl1 cell monolayers, a Nonidet P-40 gentle lysis procedure was used (21). Direct RNA sequencing was carried out by a modification of a dideoxy termination protocol (12, 32).
Revertant analysis.
Independent spontaneous revertants of mutant 3 (Alb154) and mutant 4 (Alb183) were isolated starting from individual plaques obtained at 39°C from passage 2 stocks. Each plaque was serially passaged 10 times (mutant 3) or 11 times (mutant 4) at 37°C at an estimated multiplicity of 0.2 PFU/cell. Following the final passage, revertants were isolated as viruses forming large (wild-type-size) or intermediate-size (between mutant size and wild-type size) plaques at 39°C. Mutations responsible for reversion were determined by sequencing of the E gene and other genes in RNA from cells infected with purified plaques or in purified virion RNA.
Electron microscopy.
Samples of mutant 4 (Alb183) and the isogenic wild-type strain Alb129, freshly passaged at 33°C, were harvested, clarified of cell debris by centrifugation for 10 min at 1,200 × g, and used directly for electron microscopy without further concentration or purification (see Fig. 7 and 8). In a separate experiment (see Table 2), mutant 4 and wild type were freshly passaged at 33 and at 39°C and clarified and 8-ml samples were then concentrated by centrifugation for 2 h at 151,000 × g through 1.5 ml of 50 mM Tris-maleate (pH 6.5)–1 mM EDTA–10% glycerol onto a cushion of 100% glycerol in a Beckman SW41 rotor at 4°C. Viral pellets were resuspended in 1 ml of 50 mM Tris-maleate (pH 6.5)–1 mM EDTA. Drops (40 μl) of the different viral suspensions were placed on dental wax, and previously glow-discharged, Formvar-coated copper grids were placed on the drops for 2 to 4 min. Excess liquid was removed by wicking with filter paper, and the grids were immediately washed by floating on one or two drops of 10 mM ammonium acetate. After the final wash, the grids were briefly floated on 2% sodium phosphotungstate (pH 7.0), the excess stain liquid was removed by wicking, and the samples were viewed in a Zeiss (LEO) 910 transmission electron microscope operating at 80 keV. Micrographs were recorded at a magnification of ×25,000, and the negatives were enlarged photographically.
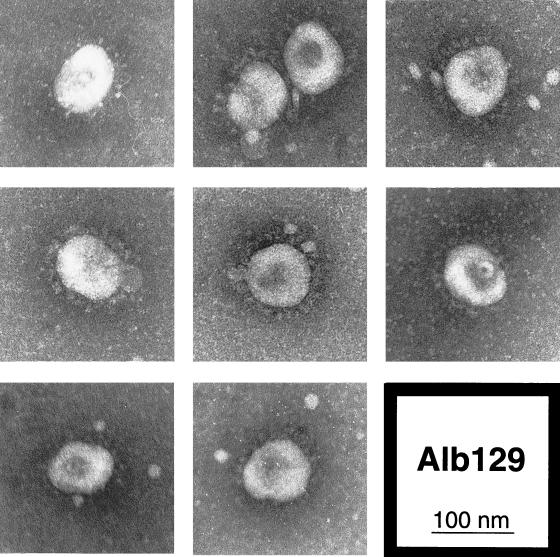
FIG. 7.
Electron microscopy of wild-type (Alb129) virions released from infected mouse 17Cl1 cells at 33°C. Freshly passaged virus was viewed by electron microscopy following negative staining with sodium phosphotungstate.
FIG. 8.
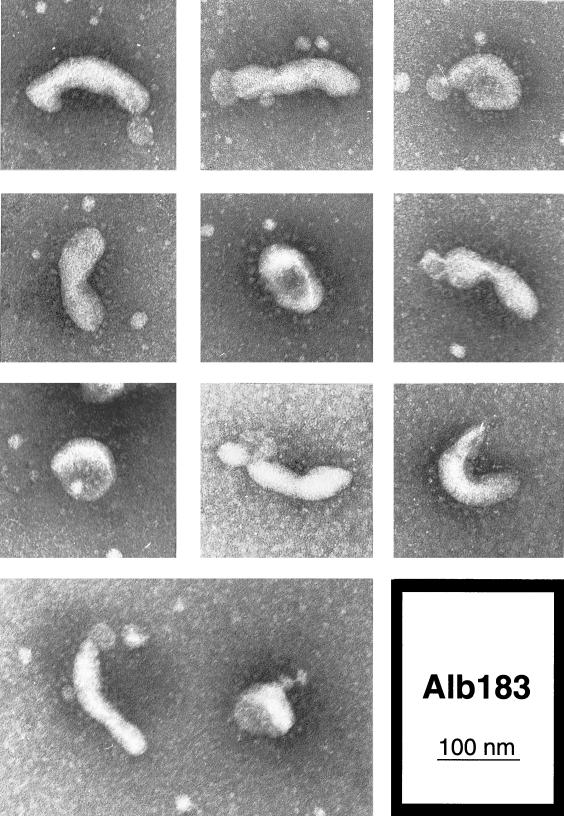
Electron microscopy of virions of E protein mutant 4 (Alb183: D60A, R61A, K63A) released from infected mouse 17Cl1 cells at 33°C. Freshly passaged virus was viewed by electron microscopy following negative staining with sodium phosphotungstate.
TABLE 2.
Virion morphology of wild type (Alb129) and E protein mutant 4 (Alb183: D60A, R61A, K63A) grown at 33 and 39°Ca
| Virus | 33°C
|
39°C
|
||
|---|---|---|---|---|
| No. (%) of normal virions | No. (%) of abnormal virions | No. (%) of normal virions | No. (%) of abnormal virions | |
| Wild type | 170 (85) | 30 (15) | 178 (89) | 22 (11) |
| Mutant 4 | 36 (18) | 164 (82) | 24 (12) | 176 (88) |
For each sample the first 200 viral particles encountered on an electron microscopy grid were scored as either normal (spherical to ovoid) or abnormal (several times longer than wide, dumbbell-shaped, or having some other noticeable structural perturbation).
Nucleotide sequence accession numbers.
The coding regions of the E genes of MHV-1, MHV-2, MHV-3, and MHV-DVIM have been assigned GenBank accession no. AF051146, AF051147, AF051148, and AF051149, respectively.
RESULTS
Comparison of MHV E protein sequences.
As a preliminary step in establishing the importance of different regions and residues of the E protein, we determined the sequences of the E genes of MHV-1, MHV-2, MHV-3, and MHV-DVIM. An alignment of the resulting deduced amino acid sequences with that of our reference strain, MHV-A59, and with those previously determined for MHV-S and MHV-JHM, was performed to obtain a sampling of the range of sequence variation allowed for the E protein (Fig. 1). This comparison presumes, and all existing interstrain recombinational evidence supports the idea, that these E proteins are functionally interchangeable.
FIG. 1.
Amino acid sequence comparison of the E proteins of seven MHV strains. The deduced MHV-1, MHV-2, MHV-3, and MHV-DVIM E sequences were determined in the present work. The E sequences of MHV-A59 (8), MHV-JHM (40), and MHV-S (31, 46) have been reported previously (GenBank accession numbers M16602, X04997, and M64835, respectively). Spaces in the alignment indicate positions at which the amino acid is identical to that of MHV-A59; dots denote every 10th residue.
As shown in Fig. 1, the MHV E proteins are all quite small, comprising 83 to 88 residues (9.6 to 10.2 kDa), and exhibit a high degree of sequence identity (89 to 98%). Almost one-third of the protein is made up of a relatively long hydrophobic region, a putative membrane anchor spanning residues V10 through I37. Although the topology of the E protein has not been unequivocally established, evidence from a study of the E protein of transmissible gastroenteritis virus (16) suggests that the molecule is in a C-exo N-endo configuration. The membrane anchor is highly conserved among the seven strains of MHV and is flanked by two invariant charged residues (D8 and K38). Four absolutely conserved cysteine residues (C23, C40, C44, and C47), which are potential sites for palmitoylation (47) or interchain disulfides, are found within the membrane anchor or adjacent to it on the carboxy side. The only hydrophilic portion of the molecule occurs near the carboxy terminus. Although the charged residues here are highly conserved, this segment represents the most divergent part of the E protein, most notably at the extreme carboxy terminus, which shows a 5-amino-acid extension (IIQTL) in many of the strains.
Generation of clustered charged-to-alanine mutants of the MHV E protein.
To address whether the E gene is an essential gene, we first attempted, by targeted RNA recombination, to create a null mutant of MHV-A59 in which the E gene was replaced by the gene for the green fluorescent protein. While we could successfully construct multiple recombinants in which gene 4 was replaced by the green fluorescent protein gene (14), we were not able to generate such a deletion in the E gene despite repeated efforts. Although this negative result could not be interpreted unequivocally, it implied that ablation of the E gene would be lethal to the virus.
We thus sought next to construct conditional lethal E gene mutants by using the strategy of clustered charged-to-alanine mutagenesis (2, 3, 44). Owing to the small size and limited hydrophilicity of the E protein, the algorithm of Wertman et al. (44) suggested only four possible sets of mutations, all of them falling in the carboxy-terminal one-third of the molecule (Fig. 2). The four clustered charged-to-alanine mutants were constructed in transcription vectors that also contained a phenotypically silent marker in gene 4 (14) and were then introduced into the MHV genome by targeted recombination. Synthetic mutant donor RNAs were independently transfected into cells infected with the N gene deletion mutant Alb4 (Fig. 3), and released progeny viruses were harvested and analyzed for the desired mutations. Since we were interested in generating temperature-sensitive mutants, plaques of progeny virus were first assayed under nonpermissive conditions (39°C), either directly or with prior heat treatment, to eliminate mutants capable of forming large (i.e., wild-type-size) plaques at this temperature. Only one of the four possible mutants, mutant 2 (designated Alb153), was found to fall into this category and its apparently wild-type phenotype was not characterized further. Although viruses forming large plaques at 39°C were identified among progeny from the other three donor RNAs, these proved to be either revertants of Alb4 or recombinants that had repaired the Alb4 deletion but did not contain the upstream E gene mutations.
Other clustered charged-to-alanine mutants had to be identified by molecular screening, because there was no other clear basis for differentiating them from the Alb4 parent, which is also temperature sensitive. Since we expected that the E mutants would be temperature sensitive but not thermolabile (or not as thermolabile as Alb4 [22]), progeny viruses from targeted recombination were heat treated (22) to enrich for potential recombinants. In retrospect, this may have hindered rather than helped subsequent screening. Pools of candidate recombinants, obtained as progeny plaques at the permissive temperature of 33°C, were assayed by RT-PCR, and individual plaques from pools that gave positive signals were then analyzed further, as described in Materials and Methods. This process yielded a recombinant of mutant 3 (Alb154) and a recombinant of mutant 4 (Alb183). A recombinant representing mutant 1 was never obtained among the numerous candidates screened, suggesting that this mutant, if not lethal, was more impaired than the Alb4 parent. Analysis of mutants 2, 3, and 4 by RT-PCR and by direct sequencing of purified genomic RNA revealed that each had the constructed set of four or five point mutations in the E gene, resulting in the conversion of two or three charged residues to alanines (Fig. 4). The sequence shown in Fig. 4 for Alb183 was obtained with considerable difficulty owing to the small amount of genomic RNA that could be purified from this virus, possibly because of virion fragility (see below). The mutations in this virus were verified in total cellular RNA isolated from Alb183-infected cells and also in revertants of Alb183. As expected, the Alb4 N gene deletion in each of the three recombinants had been repaired, and in addition, the gene 4 tag from the donor RNA was present (data not shown). This distribution of markers was consistent with each recombinant having been generated by a single crossover event upstream of gene 4 (Fig. 3).
FIG. 4.
Portions of genomic RNA sequence within the E genes of clustered charged-to-alanine mutant 2 (Alb153), mutant 3 (Alb154), and mutant 4 (Alb183). The lane markers indicate the terminating dideoxynucleoside triphosphate for each reaction. The segments of sequence show the derived positive-sense RNA sequence with its corresponding translation adjacent. Stars denote mutated nucleotides.
At elevated temperatures, the mutant 3 and mutant 4 recombinants, Alb154 and Alb183, each produced significantly smaller plaques than those of wild-type MHV-A59 or Alb129, an isogenic recombinant that contains the gene 4 tag but no other mutations (14). As indicated in Table 1, this partially temperature-sensitive phenotype was evident at both 37 and 39°C. At either of these temperatures, each mutant produced plaques that were less than one-third the diameter of wild-type plaques, although they were not as small as plaques of the N gene deletion mutant Alb4.
TABLE 1.
Plaque sizes of E mutants at different temperatures
| Virus | Mutation(s) | Plaque size (mm)a at:
|
||
|---|---|---|---|---|
| 33°C | 37°C | 39°C | ||
| Alb154 (mutant 3) | E protein K63A, K67A; gene 4 tag | 0.5 | 0.9 | 0.9 |
| Alb183 (mutant 4) | E protein D60A, R61A, K63A; gene 4 tag | 0.5 | 0.9 | 0.9 |
| Alb129 | Gene 4 tag | 0.6 | 2.8 | 3.0 |
| Wild type | None | 0.7 | 2.9 | 3.0 |
| Alb4 | N protein Δ380–408 | 0.5 | 1.0 | 0.5 |
Plaque diameters were measured at 48 h after infection of L2 cell monolayers. Each value is the average from 20 plaques.
Revertant analysis.
The sizes of plaques of mutants 3 and 4 differed sufficiently from those of plaques of the wild type to serve as the basis for the isolation of revertants. Following either 10 or 11 serial passages at the nonpermissive temperature of virus stocks originating from independent plaques of mutant 3 or mutant 4, spontaneous revertants were selected as viruses able to form plaques detectably larger than those of the mutant from which they were derived. In most cases, revertants formed wild-type-size plaques at 39°C; in some cases, the revertant plaque size was intermediate between those of the wild type and the mutant. It should be noted that despite the selective pressure applied over multiple passages, not every stock yielded a revertant.
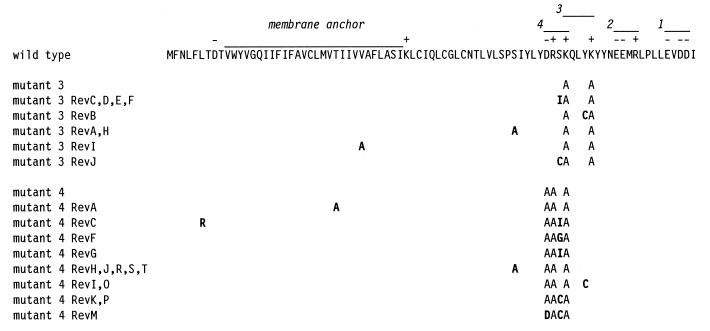
For mutant 3, nine independent revertants were isolated. Sequencing of the E genes of these revealed that each contained a single second-site mutation in the E protein and that they could be divided into five groups (Fig. 5). Similarly, 14 independent revertants of mutant 4 were isolated and were divided into eight groups (Fig. 5). In most cases, each mutant 4 revertant also contained a single second-site mutation. The two exceptions to this observation, mutant 4 RevC and mutant 4 RevM, each contained an additional mutation that was most probably of no significance. For mutant 4 RevC, the S62I mutation that it had in common with mutant 4 RevG must have been sufficient for reversion; likewise, for mutant 4 RevM, the S62C mutation that it had in common with mutant 4 RevK and mutant 4 RevP must have been sufficient for reversion.
FIG. 5.
Revertant analysis of mutant 3 (Alb154: K63A, K67A) and mutant 4 (Alb183: D60A, R61A, K63A). For each mutant or revertant, only amino acid residues differing from the wild-type MHV-A59 residue (top line) are shown; second-site mutations in the revertants are in boldface type.
Almost all of the second-site reverting mutations of both mutant 3 and mutant 4 fell at one of three positions close to the original clustered charged-to-alanine mutations in the E gene, and almost all of these (S55A, S62I, S62C, and Y66C) occurred in both revertant sets (Fig. 5). Surprisingly, the two revertants that deviated from this pattern, mutant 3 RevI and mutant 4 RevA, each had a relatively conservative amino acid substitution within the putative membrane anchor (V31A and T27A, respectively). This may suggest either that compensatory structural changes within the membrane domain of E protein can be transmitted to the ectodomain or else that monomer-monomer interactions within the membrane can make up for a loss of interaction between ectodomain moieties.
To ascertain whether the observed second-site mutations were indeed responsible for reversion, we analyzed in more detail a subset of mutant 3 revertants representative of each position at which second-site mutations were found. Initially, we sequenced the entire M gene of each member of this group, mutant 3 RevA, mutant 3 RevB, mutant 3 RevD, and mutant 3 RevI, to check for the possible presence of mutations in this protein, which potentially interacts directly with the E protein (4, 42). No mutations were found in any of the M genes of these revertants. Next, we attempted to introduce each reverting mutation, in conjunction with its cognate set of clustered charged-to-alanine mutations, into the MHV genome. This was carried out by targeted recombination between the appropriate donor RNA construct and the Alb4 N gene deletion mutant. For mutant 3 RevA, mutant 3 RevB, and mutant 3 RevD, recombinants were obtained that contained both the mutant 3 clustered charged-to-alanine mutations and the corresponding second-site mutation. Since all of these produced large plaques at 39°C, this demonstrated that the E gene second-site mutation found in each of these revertants was sufficient to compensate for the original mutations of mutant 3. However, for reasons that we currently do not understand, repeated attempts to generate a recombinant corresponding to mutant 3 RevI were unsuccessful. This result, coupled with the unexpected membrane anchor location of the second-site mutation in mutant 3 RevI, prompted us to sequence the remaining structural genes (S and N) of this virus as well as the packaging signal domain within gene 1b (15). No mutations other than that in the E protein (V31A) were found. Thus, although we cannot rule out the possibility of a compensating mutation in a nonstructural gene, it is likely that the V31A mutation accounted for the reversion of mutant 3 RevI.
Taken together these analyses confirmed that for both mutant 3 and mutant 4 one or more of the clustered charged-to-alanine mutations constructed in the E gene was responsible for the observed plaque size phenotype. Moreover, the results demonstrated that a single second-site amino acid change within the E protein was able to reverse the phenotypic effect of the original mutation(s). No intergenic suppressors of the mutant 3 or mutant 4 mutations have yet been found.
Phenotype of E protein mutants.
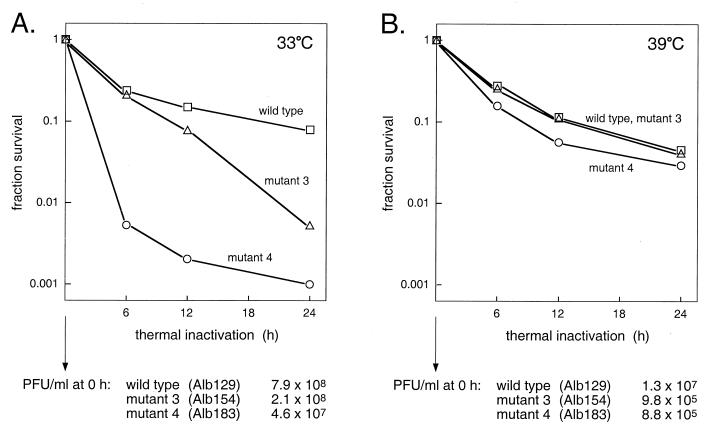
Difficulties encountered in the preparation of purified virions of mutant 3 and mutant 4 led us to suspect that there might be defects in their structural integrity. To test the thermolability of these mutants, stocks of mutant 3, mutant 4, and their isogenic wild-type counterpart, Alb129, were grown at 33°C and also at 39°C. Released viruses were then heat treated for various intervals at 40°C and pH 6.5 (22), and survivors were measured by plaque titer determination at 37°C. As shown in Fig. 6A, virions of the two mutants were at least an order of magnitude more thermolabile than were wild-type virions when grown at 33°C. By 24 h of heat treatment, the infectious titers of mutant 3 and mutant 4 had dropped 15- and 78-fold, respectively, more than that of the wild type. In contrast, when grown at 39°C, both mutant 3 and mutant 4 were much less strongly affected by heat treatment and did not differ significantly from the wild type, which showed roughly the same heat sensitivity at 39°C as at 33°C (Fig. 6B). However, the infectious titers of stocks of the two mutants grown at 39°C were markedly lower than those of stocks grown at 33°C. One explanation consistent with these data is that for the two mutants grown at 33°C, most virions were assembled in a flawed manner that was sufficient for their stability at the lower, permissive temperature but led to a loss of viability upon heat treatment. Conversely, for mutant viruses grown at 39°C, only a relatively small fraction of virions assembled successfully, but these had passed some threshold of structural stability and were no more susceptible to heat treatment than were wild-type virions. Further work is required to more precisely elucidate the basis for the thermolability differences observed in Fig. 6. It should be noted that both the plaque size and the frequency of survivors of heat treatment among mutants 3 and 4 grown at 39°C ruled out the possibility that these survivors were revertants.
FIG. 6.
Thermal inactivation of mutant 3 (Alb154: K63A, K67A), mutant 4 (Alb183: D60A, R61A, K63A), and the isogenic wild-type strain (Alb129). (A) Passage 3 stocks of each virus were grown at 33°C and thermally inactivated at 40°C (pH 6.5) for the indicated times (22). Titers of surviving viruses were determined at 37°C. (B) Same as panel A but with virus stocks grown at 39°C. The titers of virus stocks prior to heat treatment are indicated at the bottom.
For mutant 4, the more defective of the two E protein mutants, we next sought further indications of structural impairment. When virions of mutant 4 were examined by electron microscopy, a remarkable difference from virions of the isogenic wild-type control, Alb129, was seen. Negatively stained preparations of wild type had an appearance typical for MHV and other coronaviruses (Fig. 7). With few exceptions, each virion was a roughly spherical particle 80 to 100 nm in diameter, with a more densely staining center and a complete halo of surface spikes projecting approximately 20 nm beyond the periphery of the particle. By contrast, although some virions of mutant 4 conformed to this description, most particles of this E protein mutant deviated dramatically from the typical coronavirus structure, as exemplified in Fig. 8. The majority of mutant 4 virions exhibited a more narrow, tubular appearance. These elongated virions were often pinched at multiple points, producing dumbbell-shaped forms. Even the more spherical particles of mutant 4 frequently contained two or three lobes. Between 80 and 90% of mutant 4 virions, whether grown at 33 or at 39°C, could be classified as having an abnormal morphology, whereas no more than 15% of wild-type virions fell into this category (Table 2). The estimated volumes of mutant 4 virions were, on average, similar to those of wild-type virions but varied much more from the mean value. As can be seen in Fig. 8, despite the abnormality of virion shape, the mutant showed no detectable aberration in its distribution of surface spikes. These observations provided striking evidence for the critical role of the coronavirus E protein in virion morphogenesis, clearly corroborating the results obtained by expression studies that established the requirements for virus-like particle formation (4, 42).
DISCUSSION
In this study, we have constructed the first coronavirus E gene mutants used then to show that the E protein of MHV is fundamental to the correct assembly of virions. Initially, we determined the sequences of the E proteins of a number of strains of MHV. This added to the accumulated evidence that, even though deletion, frameshift, or transcriptionally inactive MHV mutants have been identified in the HE gene (28), gene 2 (37), gene 4 (31, 41, 43, 46), and gene 5a (31, 46), the E gene is always found to be intact. Furthermore, comparison of our data with other available MHV E sequences showed that this protein is highly conserved. Most of the relatively limited set of divergent residues tend to be grouped in the hydrophilic carboxy-terminal one-third of the molecule. Within this region, however, charged amino acids almost always remain invariant. Heterogeneity also occurs near the carboxy terminus of the E protein in other coronavirus species for which more than one sequence is known. In particular, it has been noted that this portion of the avian infectious bronchitis virus E protein (the product of gene 3c) not only shows primary sequence differences among a number of strains but also varies in length by as many as 16 residues (25).
The overall conservation of the MHV E protein may be taken to argue that the E protein is critical, if not essential, for the virus. Thus, it is probably not remarkable that we were unable to construct a deletion mutant in which E would have been replaced with a heterologous gene. However, the limitations of targeted recombination do not allow us to draw a definitive conclusion from this negative result, and it is conceivable that complete removal of the E gene might not be lethal. The gene for the 6K protein of Semliki Forest virus, which in some respects may be analogous to E, can be deleted to produce mutants that are severely impaired but still viable (24, 27). It will be possible to more decisively test whether the E gene is absolutely essential if an infectious cDNA of the MHV genome becomes available.
The conservation of charged residues in the MHV E molecule supported the underlying logic of our effort to seek conditional lethal E gene mutants by the strategy of clustered charged-to-alanine mutagenesis. Of the four possible mutants that we attempted to create, we were able to obtain three, and two of these exhibited an impaired phenotype relative to wild-type MHV. These two, mutant 3 (Alb154) and mutant 4 (Alb183), are the first mutants we have constructed by targeted RNA recombination that were identified by screening rather than by selection, and they thus represent a broadening of the range and usefulness of this method for genetic studies. Numerous independent, spontaneous revertants of the two E mutants were isolated and analyzed. As expected, since the original mutants contained multiple mutations, all the revertants resulted from second-site mutations. Each was found to contain a single relevant reverting mutation in the proximity of the original E gene clustered charged-to-alanine mutations. We had hoped that some of the second-site revertants would map elsewhere than in the E gene and thus provide evidence for sites of interaction between E and another viral protein, but we have not yet found a revertant of this type. Based on the studies of Vennema et al. (42) and Bos et al. (4), we favor the notion of direct intermolecular contacts between E protein and M protein, but it is possible that E protein can bring about virion budding without such an association. Alternatively, the lesions in mutant 3 and mutant 4 may be such that they cannot be repaired by a compensating change in any protein other than E, while other portions of the molecule may interact with the M protein.
The localization of all reverting mutations within the E gene strongly indicated that the phenotypes of mutant 3 and mutant 4 were indeed caused by the mutations (or a subset of the mutations) originally constructed in these recombinants and were not due to hypothetical secondary mutations that could have arisen during their isolation. This conclusion was reinforced by the demonstration for a subset of mutant 3 revertants that reintroduction of a reverting mutation in conjunction with its cognate set of clustered charged-to-alanine mutations resulted in a wild-type phenotype. Taken together, these results show that at least one hydrophilic region outside the membrane can play an important role in E protein function, but a role for oligomeric associations between membrane anchor regions may also be inferred by the surprising loci of two of the reverting mutations: V31A (in mutant 3 RevI) and T27A (in mutant 4 RevA). Although these changes are relatively small, their significance may be that threonine and valine are more rigid and bulky than alanine. Hence, the mutation to alanine may increase the flexibility of the membrane domain to allow an adjustment in the contact with the membrane domain of another E monomer that compensates for the destabilization introduced by the clustered charged-to-alanine mutations in the ectodomain. Alternatively, we can hypothesize that the residues V31 and T27 are not actually located within the membrane domain but just downstream of it and that they directly interact with the carboxy terminus of the E protein. This would suggest that the membrane anchor of the E protein is 17 rather than 28 amino acids, consistent with the fact that transmembrane segments of Golgi proteins are generally shorter than those of plasma membrane proteins and that 15 residues are sufficient to cross the Golgi membrane (6). Considerably more information about the E protein is required before we can understand the structural basis for the action of all of the reverting mutations.
The phenotypic properties of the E mutants, especially mutant 4, plainly substantiate the importance of the MHV E protein in virion morphogenesis. The precise role of E in the formation of the viral particle, however, remains to be clearly defined. The thermolability data of Fig. 6 do not enable us to distinguish between E as an active structural constituent of the membrane envelope or as an accidental passenger that appears in the budded virion but has already performed its function at a prior step of assembly. For virions assembled at 33°C, the mutant E protein may adopt a defective conformation upon heating. On the other hand, the mutant E protein may have already acted in a defective manner establishing improper associations among M protein monomers that disrupt virions upon elevation of temperature. Likewise, the electron micrographs of mutant 4 (Fig. 8) revealed a distinct contrast with wild-type virions (Fig. 7), but further experiments remain to establish the events that distorted the assembly process for the mutant. Pertinent to the range of sizes of the mutant particles, we do not yet know if every aberrant particle contained a nucleocapsid or if every particle contained only one nucleocapsid. Many of the mutants of the 6K protein of Sindbis virus have been found to package multiple nucleocapsids within single membrane envelopes (20). One potential role that has been proposed for the coronavirus E protein is that it is the factor that pinches off the “neck” of the nascent virion particle in the final stages of budding (42). We might speculate, then, that the multiple pinches observed in many of the mutant 4 particles (Fig. 8) could be examples of such a function acting in inappropriate places. We hope that additional genetic studies, complemented by protein expression studies (4, 10, 42), will establish a basis to evaluate the mode of action of E protein and approach the outstanding questions on the membrane orientation and state of oligomerization of this molecule, its effect on the budding compartment, and its possible interactions with other viral and cellular proteins.
ACKNOWLEDGMENTS
We are grateful to Cheri Koetzner for expert technical assistance and to Patrick van Roey for helpful discussion of the revertant data. We thank Tim Moran and Matthew Shudt of the Molecular Genetics Core Facility of the Wadsworth Center for the synthesis of oligonucleotides and for automated DNA sequencing. We thank Kathryn Holmes and Ehud Lavi for providing MHV strains. All electron microscopy was performed at the Electron Microscopy Core Facility of the Wadsworth Center.
This work was supported in part by Public Health Service grant AI 39544 from the National Institutes of Health.
REFERENCES
- 1.Abraham S, Kienzle T E, Lapps W E, Brian D A. Sequence and expression analysis of potential nonstructural proteins of 4.9, 4.8, 12.7, and 9.5 kDa encoded between the spike and membrane protein genes of the bovine coronavirus. Virology. 1990;177:488–495. doi: 10.1016/0042-6822(90)90513-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass S H, Mulkerrin M G, Wells J A. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc Natl Acad Sci USA. 1991;88:4498–4502. doi: 10.1073/pnas.88.10.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett W F, Paoni N F, Keyt B A, Botstein D, Jones A J, Presta L, Wurm F M, Zoller M J. High resolution analysis of functional determinants on human tissue-type plasminogen activator. J Biol Chem. 1991;266:5191–5201. [PubMed] [Google Scholar]
- 4.Bos E C W, Luytjes W, van der Meulen H, Koerten H K, Spaan W J M. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boursnell M E G, Binns M M, Brown T D K. Sequencing of coronavirus IBV genomic RNA: three open reading frames in the 5′ ‘unique’ region of mRNA D. J Gen Virol. 1985;66:2253–2258. doi: 10.1099/0022-1317-66-10-2253. [DOI] [PubMed] [Google Scholar]
- 6.Bretscher M S, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 7.Brian D A, Hogue B G, Kienzle T E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 165–179. [Google Scholar]
- 8.Budzilowicz C J, Weiss S R. In vitro synthesis of two polypeptides from a nonstructural gene of coronavirus mouse hepatitis virus strain A59. Virology. 1987;157:509–515. doi: 10.1016/0042-6822(87)90293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Décimo D, Philippe H, Hadchouel M, Tardieu M, Meunier-Rotival M. The gene encoding the nucleocapsid protein: sequence analysis in murine hepatitis virus type 3 and evolution in Coronaviridae. Arch Virol. 1993;130:279–288. doi: 10.1007/BF01309660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan C A M, Kuo L, Masters P S, Vennema H, Rottier P J M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond S E, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fichot O, Girard M. An improved method for sequencing of RNA templates. Nucleic Acids Res. 1990;18:6162. doi: 10.1093/nar/18.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer F, Peng D, Hingley S T, Weiss S R, Masters P S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer F, Stegen C F, Koetzner C A, Masters P S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godet M, L’haridon R, Vautherot J-F, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassett D E, Condit R C. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc Natl Acad Sci USA. 1994;91:4554–4558. doi: 10.1073/pnas.91.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis, a practical approach. New York, N.Y: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 19.Hsue B, Masters P S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova L, Lustig S, Schlesinger M J. A pseudo-revertant of a Sindbis virus 6K protein mutant, which corrects for aberrant particle formation, contains two new mutations that map to the ectodomain of the E2 glycoprotein. Virology. 1995;206:1027–1034. doi: 10.1006/viro.1995.1025. [DOI] [PubMed] [Google Scholar]
- 21.Kingsman S M, Samuel C E. Mechanism of interferon action. Interferon-mediated inhibition of simian virus-40 early RNA accumulation. Virology. 1980;101:458–465. doi: 10.1016/0042-6822(80)90459-6. [DOI] [PubMed] [Google Scholar]
- 22.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leibowitz J L, Perlman S, Weinstock G, DeVries J R, Budzilowicz C, Weissemann J M, Weiss S R. Detection of a murine coronavirus nonstructural protein encoded in a downstream open reading frame. Virology. 1988;164:156–164. doi: 10.1016/0042-6822(88)90631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu D X, Cavanagh D, Green P, Inglis S C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D X, Inglis S C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loewy A, Smyth J, von Bonsdorff C-H, Liljeström P, Schlesinger M J. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol. 1995;69:469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luytjes W, Bredenbeek P J, Noten A F H, Horzinek M C, Spaan W J M. Sequence of mouse hepatitis virus A59 mRNA2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker M M, Masters P S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker, M. M., and P. S. Masters. Unpublished data.
- 32.Peng D, Koetzner C A, Masters P S. Analysis of second-site revertants of a murine coronavirus nucleocapsid protein deletion mutant and construction of nucleocapsid protein mutants by targeted RNA recombination. J Virol. 1995;69:3449–3457. doi: 10.1128/jvi.69.6.3449-3457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng D, Koetzner C A, McMahon T, Zhu Y, Masters P S. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J Virol. 1995;69:5475–5484. doi: 10.1128/jvi.69.9.5475-5484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reijo R A, Cooper E M, Beagle G J, Huffaker T C. Systematic mutational analysis of the yeast beta-tubulin gene. Mol Biol Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz B, Routledge E, Siddell S G. Murine nonstructural protein ns2 is not essential for virus replication in transformed cells. J Virol. 1990;64:4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senanayake S D, Hofmann M A, Maki J L, Brian D A. The nucleocapsid protein gene of bovine coronavirus is bicistronic. J Virol. 1992;66:5277–5283. doi: 10.1128/jvi.66.9.5277-5283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddell S G. The Coronaviridae: an introduction. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–10. [Google Scholar]
- 40.Skinner M A, Ebner D, Siddell S G. Coronavirus MHV-JHM mRNA 5 has a sequence arrangement which potentially allows translation of a second, downstream open reading frame. J Gen Virol. 1985;66:581–592. doi: 10.1099/0022-1317-66-3-581. [DOI] [PubMed] [Google Scholar]
- 41.Skinner M A, Siddell S G. Coding sequence of coronavirus MHV-JHM mRNA4. J Gen Virol. 1985;66:593–596. doi: 10.1099/0022-1317-66-3-593. [DOI] [PubMed] [Google Scholar]
- 42.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss S R, Zoltick P W, Leibowitz J L. The ns 4 gene of mouse hepatitis virus (MHV), strain A59 contains two ORFs and thus differs from ns 4 of the JHM and S strains. Arch Virol. 1993;129:301–309. doi: 10.1007/BF01316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang W, Cuconati A, Paul A V, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 46.Yokomori K, Lai M M C. Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication. J Virol. 1991;65:5605–5608. doi: 10.1128/jvi.65.10.5605-5608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]