Status: In production use at Janelia. See wiki page for outside lab use of DVID.
There is a paper on DVID describing its motivation and architecture, including how versioning works at the key-value level.
See the DVID Wiki for more information including installation and examples of use.
DVID is a distributed, versioned, image-oriented dataservice written to support Janelia Farm Research Center's brain imaging, analysis and visualization efforts. It's goal is to provide:
- Easily extensible data types that allow tailoring of access speeds, storage space, and APIs.
- The ability to use a variety of storage systems by either creating a data type for that system or using a storage engine, currently limited to ordered key/value databases.
- A framework for thinking of distribution and versioning of data similar to distributed version control systems like git.
- A stable science-driven API that can be implemented either by native DVID data types and storage engines or by proxying to other connectomics services like Google BrainMaps, BOSS, etc.
DVID aspires to be a "github for large image-oriented data" because each DVID server can manage multiple repositories, each of which contains an image-oriented repo with related data like an image volume, labels, annotations, and skeletons. The goal is to provide scientists with a github-like web client + server that can push/pull data to a collaborator's DVID server.
Although DVID is easily extensible by adding custom data types, each of which fulfill a minimal interface (e.g., HTTP request handling), DVID's initial focus is on efficiently handling data essential for Janelia's connectomics research:
- image and 64-bit label 3d volumes, including multiscale support
- 2d images in XY, XZ, YZ, and arbitrary orientation
- multiscale 2d images in XY, XZ, and YZ, similar to quadtrees
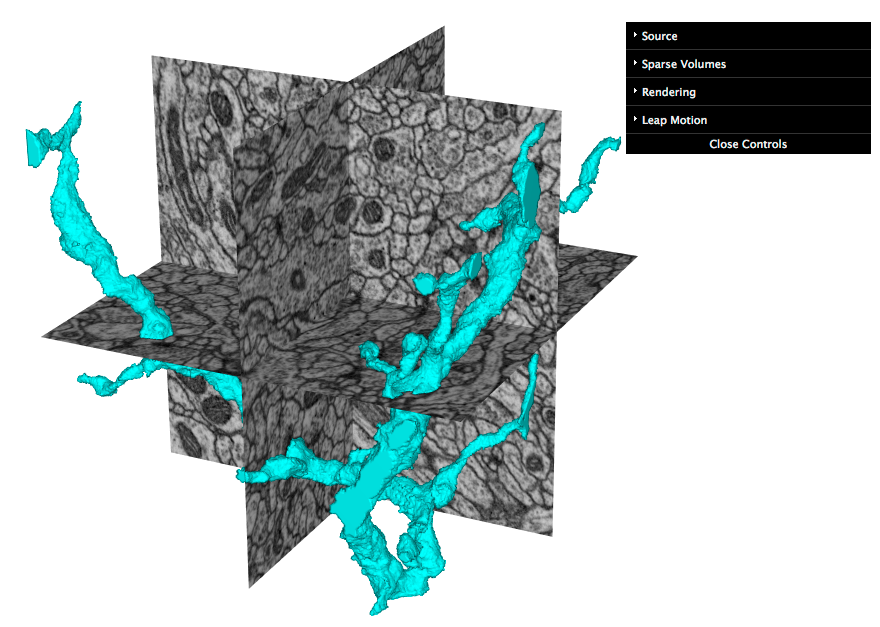
- sparse volumes, corresponding to each unique label in a volume, that can be merged or split
- point annotations (e.g., synapse elements) that can be quickly accessed via subvolumes or labels
- label graphs
- regions of interest represented via a coarse subdivision of space using block indices
- 2d and 3d image and label data using Google BrainMaps API and other cloud-based services
Each of the above is handled by built-in data types via a Level 2 REST HTTP API implemented by Go language packages within the datatype directory. When dealing with novel data, we typically use the generic keyvalue datatype and store JSON-encoded or binary data until we understand the desired access patterns and API. When we outgrow the keyvalue type's GET, POST, and DELETE operations, we create a custom datatype package with a specialized HTTP API.
DVID allows you to assign different storage systems to data instances within a single repo, which allows great flexibility in optimizing storage for particular use cases. For example, easily compressed label data can be store in fast, expensive SSDs while larger, immutable grayscale image data can be stored in petabyte-scale read-optimized systems.
DVID is written in Go and supports different storage backends, a REST HTTP API, and command-line access (likely minimized in near future). Some components written in C, e.g., storage engines like Leveldb and fast codecs like lz4, are embedded or linked as a library.
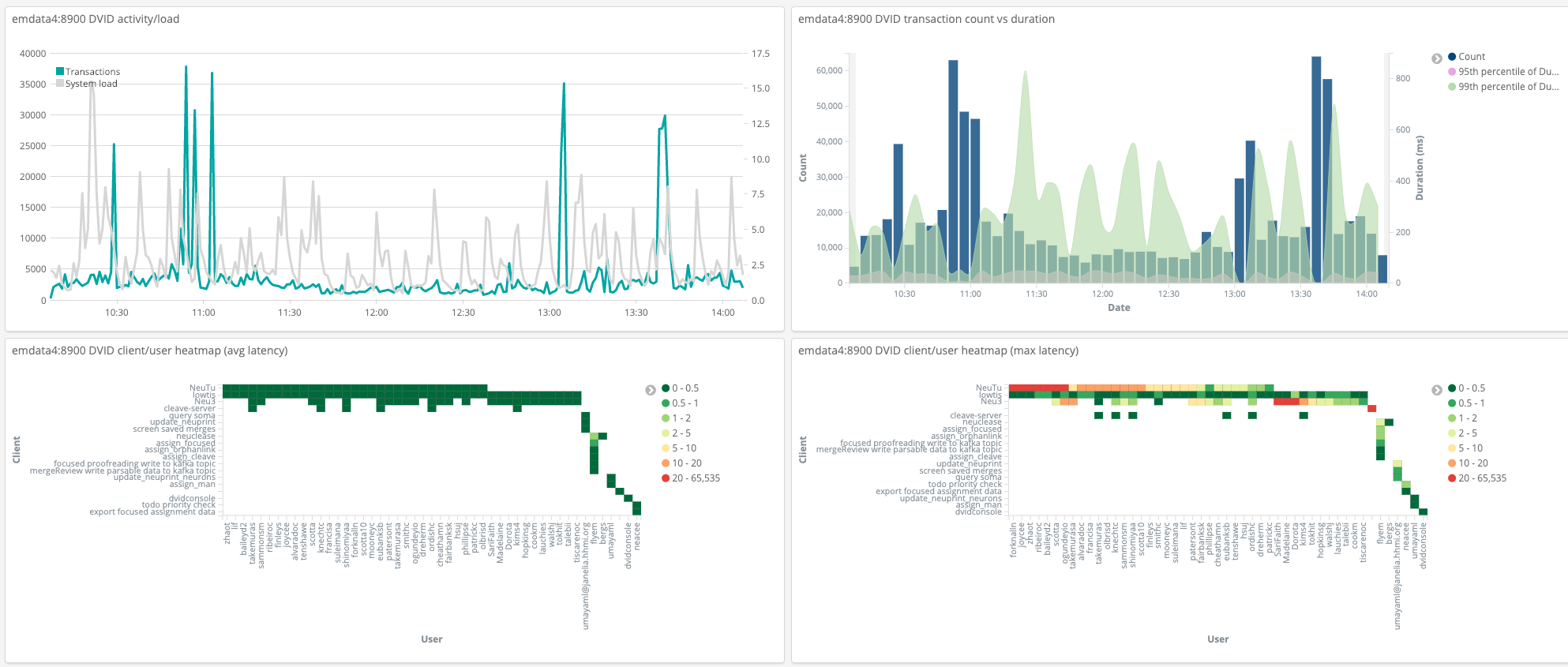
Mutations and activity logging can be sent to a Kafka server. We use kafka activity topics to feed Kibana for analyzing DVID performance.
DVID has been tested on MacOS X, Linux (Fedora 16, CentOS 6, Ubuntu), and Windows 10+ Bash Shell. It comes out-of-the-box with an embedded leveldb for storage although you can configure other storage backends.
Command-line and HTTP API documentation can be found in help constants within packages or by visiting the /api/help HTTP endpoint on a running DVID server.