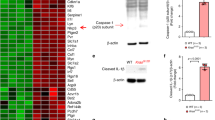
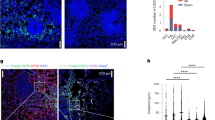
Abstract
The inflammasome plays multifaceted roles in cancer, but less is known about its function during premalignancy upon initial cell transformation. We report a homeostatic function of the inflammasome in suppressing malignant transformation through Ras inhibition. We identified increased hematopoietic stem cell (HSC) proliferation within the bone marrow of inflammasome-deficient mice. HSCs within an inflammasome-deficient stroma expressed a Ras signature associated with increased Ras pathway- and cancer-related transcripts and heightened levels of cytokine, chemokine and growth factor receptors. Stromal inflammasome deficiency established a poised Ras-dependent mitogenic state within HSCs, which fueled progeny B cell lymphomagenesis upon Myc deregulation in a spontaneous model of B cell lymphoma, and shortened its premalignant stage leading to faster onset of malignancy. Thus, the stromal inflammasome preserves tissue balance by restraining Ras to disrupt the most common oncogenic Myc–Ras cooperation and establish a natural defense against transition to malignancy. These findings should inform preventative therapies against hematological malignancies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Source data are provided with this paper.
References
Srivastava, S., Ghosh, S., Kagan, J., Mazurchuk, R. & National Cancer Institute’s HTAN Implementation. The making of a precancer atlas: promises, challenges, and opportunities. Trends Cancer 4, 523–536 (2018).
Barnett, K. C., Li, S., Liang, K. & Ting, J. P. A 360 degrees view of the inflammasome: mechanisms of activation, cell death, and diseases. Cell 186, 2288–2312 (2023).
Karki, R. & Kanneganti, T. D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 19, 197–214 (2019).
Kent, A. & Blander, J. M. Nod-like receptors: key molecular switches in the conundrum of cancer. Front. Immunol. 5, 185 (2014).
Basiorka, A. A. et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 128, 2960–2975 (2016).
Masters, S. L. et al. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity 37, 1009–1023 (2012).
Zhao, X. et al. NLRP3 inflammasome activation plays a carcinogenic role through effector cytokine IL-18 in lymphoma. Oncotarget 8, 108571–108583 (2017).
Lu, F. et al. NLRP3 inflammasome upregulates PD-L1 expression and contributes to immune suppression in lymphoma. Cancer Lett. 497, 178–189 (2021).
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985).
Harris, A. W. et al. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J. Exp. Med. 167, 353–371 (1988).
Kortlever, R. M. et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171, 1301–1315.e1314 (2017).
Land, H., Parada, L. F. & Weinberg, R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304, 596–602 (1983).
Mahauad-Fernandez, W. D. & Felsher, D. W. The Myc and Ras partnership in cancer: indistinguishable alliance or contextual relationship? Cancer Res. 80, 3799–3802 (2020).
Winkler, R., Piskor, E. M. & Kosan, C. Lessons from using genetically engineered mouse models of MYC-induced lymphoma. Cells 12, 37 (2022).
Langdon, W. Y., Harris, A. W., Cory, S. & Adams, J. M. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell 47, 11–18 (1986).
Croxford, J. L. et al. ATM-dependent spontaneous regression of early Emu-myc-induced murine B-cell leukemia depends on natural killer and T cells. Blood 121, 2512–2521 (2013).
Cervenak, L., Magyar, A., Boja, R. & Laszlo, G. Differential expression of GL7 activation antigen on bone marrow B cell subpopulations and peripheral B cells. Immunol. Lett. 78, 89–96 (2001).
Kumar, R., Godavarthy, P. S. & Krause, D. S. The bone marrow microenvironment in health and disease at a glance. J. Cell Sci. 131, jcs201707 (2018).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Kotecha, N., Krutzik, P. O. & Irish, J. M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 10, 17 (2010).
Sidman, C. L., Shaffer, D. J., Jacobsen, K., Vargas, S. R. & Osmond, D. G. Cell populations during tumorigenesis in Eu-myc transgenic mice. Leukemia 7, 887–895 (1993).
Heng, T. S., Painter, M. W. & Immunological Genome Project, C. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Amir el, A. D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Pietras, E. M. et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 (2016).
Rao, T. N. et al. JAK2-mutant hematopoietic cells display metabolic alterations that can be targeted to treat myeloproliferative neoplasms. Blood 134, 1832–1846 (2019).
Howard, J. E., Smith, J. N. P., Fredman, G. & MacNamara, K. C. IL-18R-mediated HSC quiescence and MLKL-dependent cell death limit hematopoiesis during infection-induced shock. Stem Cell Reports 16, 2887–2899 (2021).
Silberstein, L. et al. Proximity-based differential single-cell analysis of the niche to identify stem/progenitor cell regulators. Cell Stem Cell 19, 530–543 (2016).
Ivanova, N. B. et al. A stem cell molecular signature. Science 298, 601–604 (2002).
Eppert, K. et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17, 1086–1093 (2011).
Gazit, R. et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 211, 1315–1331 (2014).
Antico Arciuch, V. G., Elguero, M. E., Poderoso, J. J. & Carreras, M. C. Mitochondrial regulation of cell cycle and proliferation. Antioxid. Redox Signal. 16, 1150–1180 (2012).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Satoh, T., Fantl, W. J., Escobedo, J. A., Williams, L. T. & Kaziro, Y. Platelet-derived growth factor receptor mediates activation of ras through different signaling pathways in different cell types. Mol. Cell. Biol. 13, 3706–3713 (1993).
Weijzen, S. et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8, 979–986 (2002).
Baumgart, A. et al. Opposing role of Notch1 and Notch2 in a Kras(G12D)-driven murine non-small cell lung cancer model. Oncogene 34, 578–588 (2015).
Klomp, J. E., Klomp, J. A. & Der, C. J. The ERK mitogen-activated protein kinase signaling network: the final frontier in RAS signal transduction. Biochem. Soc. Trans. 49, 253–267 (2021).
Tikhonova, A. N. et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228 (2019).
Morrison, S. J. & Scadden, D. T. The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014).
Wei, Q. & Frenette, P. S. Niches for hematopoietic stem cells and their progeny. Immunity 48, 632–648 (2018).
Ding, L. & Morrison, S. J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013).
Zhu, J. et al. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood 109, 3706–3712 (2007).
Lieberman, J., Wu, H. & Kagan, J. C. Gasdermin D activity in inflammation and host defense. Sci. Immunol. 4, eaav1447 (2019).
Hoggatt, J., Kfoury, Y. & Scadden, D. T. Hematopoietic stem cell niche in health and disease. Annu. Rev. Pathol. 11, 555–581 (2016).
Comazzetto, S., Shen, B. & Morrison, S. J. Niches that regulate stem cells and hematopoiesis in adult bone marrow. Dev. Cell 56, 1848–1860 (2021).
Fernandez-Medarde, A., De Las Rivas, J. & Santos, E. 40 years of RAS—a historic overview. Genes (Basel) 12, 681 (2021).
Prior, I. A., Hood, F. E. & Hartley, J. L. The frequency of Ras mutations in cancer. Cancer Res. 80, 2969–2974 (2020).
Zhou, B., Der, C. J. & Cox, A. D. The role of wild type RAS isoforms in cancer. Semin. Cell Dev. Biol. 58, 60–69 (2016).
Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 (2019).
Meizlish, M. L., Franklin, R. A., Zhou, X. & Medzhitov, R. Tissue homeostasis and inflammation. Annu. Rev. Immunol. 39, 557–581 (2021).
Park, S. S. et al. Insertion of c-Myc into Igh induces B-cell and plasma-cell neoplasms in mice. Cancer Res. 65, 1306–1315 (2005).
Ullman-Cullere, M. H. & Foltz, C. J. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab. Anim. Sci. 49, 319–323 (1999).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Reiner, A., Yekutieli, D. & Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 (2003).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Zappia, L. & Oshlack, A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience 7, giy083 (2018).
McCarty, E. et al. Single cell transcriptomics of bone marrow derived macrophages reveals Ccl5 as a biomarker of direct IFNAR-independent responses to DNA sensing. Front. Immunol. 14, 1199730 (2023).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Jerome, A. D. et al. Cytokine polarized, alternatively activated bone marrow neutrophils drive axon regeneration. Nat. Immunol. 25, 957–968 (2024).
Shen, C. et al. Inflammasome protein scaffolds the DNA damage complex during tumor development. Nat. Immunol. 25, 2085–2096 (2024).
Gamez-Garcia, A. et al. A SIRT7-dependent acetylation switch regulates early B cell differentiation and lineage commitment through Pax5. Nat. Immunol. 25, 2308–2319 (2024).
Markowitz, G. J. et al. Deficiency of metabolic regulator PKM2 activates the pentose phosphate pathway and generates TCF1+ progenitor CD8+ T cells to improve immunotherapy. Nat. Immunol. 25, 1884–1899 (2024).
He, K. et al. Spatial microniches of IL-2 combine with IL-10 to drive lung migratory TH2 cells in response to inhaled allergen. Nat. Immunol. 25, 2124–2139 (2024).
Acknowledgements
This work was supported by institutional seed funds to J.M.B., grant no. F30CA174313 to A.K. and training grants no. T32AI007605 to A.K. and no. T32DK116970 to K.J.Y.M. J.M.B. and the Blander Lab are also supported by National Institutes of Health grants to Principal Investigator J.M.B. (grant nos. AI170832, AI170897, AI159772, AI178327), and the Binational US–Israel Science Foundation Grant no. 2021299. G.B. was supported by fellowship and career development awards from the Crohn’s & Colitis Foundation and the Robert Wood Johnson Foundation Grant no. 74260. G.I.E. is supported by a Cancer Research UK Program Award no. A29210 (Principal Investigator G.I.E.).
Author information
Authors and Affiliations
Contributions
A.K., K.J.Y.M. and J.M.B. designed the experiments. A.K. and K.J.Y.M. performed the experiments. A.K., K.J.Y.M. and J.M.B. wrote the manuscript and prepared figures. A.K. and J.M.B. directed the study. B.J. and G.B. sorted HSCs and prepared samples for RNA-seq. G.P. and Z.H. performed RNA-seq data processing and expression analyses. D.Z. performed the cytokine arrays. A.G. performed bioinformatic analyses of published bone marrow stroma scRNA-seq datasets, and A.G. and J.M.B. designed Seurat cluster analysis and cell type annotation. R.M.K. and G.I.E. contributed to the design of cytokine array and Ras inhibition-related experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Gabriel Mbalaviele, James Vince and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in colloboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analyses of GL-7 and IgM expression facilitate characterization of premalignant and malignant Eμ-myc B cells in circulation.
a. Flow cytometry gating on CD19+ B cells from the peripheral blood of representative WT and Eμ-myc mice, at 4 weeks of age, depicting GL-7 and B220 staining within the IgM+ and IgM− fractions of the circulating CD19+ population of B cells. Data represent hundreds of mice genotyped using this gating strategy. b,c, Flow cytometry on and frequencies of circulating pre-B cells at 4, 6, and 12 weeks of age (n ≥ 4 per group). d, Diversity of malignant CD19+ cellular phenotypes. Each plot depicts the circulating CD19+ cells from an individual mouse at the malignant stage of disease. e, Representative Ly6C and F4/80 staining patterns within whole bone marrow. f, Representative conventional flow cytometry gating strategy to delineate B cells, T cells, and neutrophils. Data represent four independent experiments, at least four mice per genotype. g, Absolute numbers of T cells, neutrophils, monocytes and macrophages as defined in (f) from the bone marrow of mice of each genotype, and calculated ratio of macrophages to monocytes (bottom panel). Each dot represents an individual mouse (n ≥ 4 per genotype). h, cytometric bead array quantification of cytokines from bone marrows of mice of the indicated genotypes. Each dot represents an individual mouse (n ≥ 8 per genotype). *=p ≤ 0.05, **=p ≤ 0.01, ***=p ≤ 0.001, calculated by two-tailed unpaired student’s T-test. All quantified data are presented as mean values +/− SEM.
Extended Data Fig. 2 CyTOF visualization of hematopoietic cell lineages and cell type specific expression of Casp1 and Casp11.
a, Representative SPADE tree analysis of CyTOF data depicting the major hematopoietic stem cell, progenitor, and committed lineage clusters from the mouse bone marrow. b, SPADE tree diagrams from each of the Eu-myc and Casp1−⁄− Casp11−⁄− mouse genotypes showing increased frequency in the B cell progenitor cluster in Eu-myc Casp1−⁄− Casp11−⁄− mouse bone marrow. c,d, Average Casp1 (c) and Casp11 (d) relative expression values, in arbitrary units (AU) extracted for each indicated cell population from the ImmGen database (SC.LT34f.BM = Long-term reconstituting stem cell; SC.LTSL.BM = long-term repopulating hematopoietic stem cell; SC.STSL.BM = short-term repopulating hematopoietic stem cell; SC.MPP34F.BM = Flk2+ multipotent progenitor; SC.ST34F.BM = Flk2− multipotent progenitor; SC.CMP.BM.MM = common myeloid progenitor; SC.MEP.BM = megakaryocyte erythroid progenitor; SC.GMP.BM = granulocyte-monocyte progenitor; SC.CDP.BM = common dendritic cell precursors; SC.MDP.BM = monocyte-dendritic cell precursors; MLP.BM = multilineage progenitor; proB.CLP.BM = common-lymphoid progenitor; proB.FrA.BM = prepro-B cells; preB.FrBC.BM = pro-B cells; preB.FrC.BM = cycling pre-B cells; preB.FrD.BM = small pre-B cells; B.FrE.BM = newly-formed B cells; B.FrF.BM = Bone marrow fraction F B cells). e, Sorting strategy for representative LSK, B cell, macrophage, and monocyte cell populations. f, Relative Casp1 expression levels from each sorted population from (e) as determined by qRT-PCR. g, Relative Casp11 expression levels from each sorted population from (e) as determined by qRT-PCR. Data in e-g represent two independent experiments, totaling 4 biological replicates. *=p ≤ 0.05, **=p ≤ 0.01, ***=p ≤ 0.001, calculated by two-tailed unpaired student’s T-test. All quantified data are presented as mean values +/− SEM.
Extended Data Fig. 3 Inflammasome impairment alters the frequency but not phenotype of premalignant B cell progenitors.
a, Wright staining of MACS-isolated CD19+ cells from the bone marrow of indicated genotypes. Data is representative of 2 independent experiments with n = 3 total biological replicates/genotype. b, Representative forward scatter (FSC-A) of CD19+ and GL-7+IgM− cells from the indicated genotypes. c, CYTOF viSNE plots of representative bone marrow samples of each genotype with colors indicating cells within the indicated marker gates. d, viSNE cell density diagrams on the same samples as in (c). Data represent three independent experiments with six individual mice total per genotype. e, Representative B cell marker expression from the CyTOF data, comparing genotypes. Data represent three independent experiments with six individual mice total per genotype. f, Intracellular cleaved caspase-3 (CC3) vs. GL-7 staining within the CD19+ compartment. g,h, Quantification of (f) showing % CC3+ in CD19+ gate (g) and the CD19+GL-7+ gate (h) (n ≥ 4 mice per group). All experiments were performed on 4-week-old mice of the indicated genotypes. *=p ≤ 0.05, **=p ≤ 0.01, ***=p ≤ 0.001, calculated by two-tailed unpaired student’s T-test. All quantified data are presented as mean values +/− SEM.
Extended Data Fig. 4 FACS-sorted HSCs are highly enriched for HSC-specific genes.
a, Flow cytometry contour plots showing gating on lineage−Sca-1+c−-Kit+ (LSK) stem cells prior to FACS-sorting of cells for RNA-sequencing. LSK cells were isolated from total bone marrow cells extracted from femurs and tibias of indicated premalignant 6-week-old mice and stained for specific LSK cells isolation with an antibody mixture containing the Lineage marker cocktail (clones: 145-2C11, RB6-8C5, M1/70, RA3-6B2, Ter-119), anti-c-Kit (clone 2B8) and anti-Sca-1 (clone D7) antibodies. b,c, Histogram of mean normalized counts of sequenced genes from sorted LSK cells showing high enrichment of genes restricted to HSC as reported by the laboratories of Lemischka and Dick29,30, compared to relatively lower enrichment of HSC genes reported by Rossi laboratory31 (b), and highest enrichment compared to signature HSC genes represented in the Hematopoietic Stem Cell Differentiation gene ontology biological process (b,c). Positioning of the average total counts of genes in our sorted LSK HSC cells is indicated as ‘Total’.
Extended Data Fig. 5 Distinct bone marrow profiles of caspase 1 and 11 deficient mice irrespective of Eμ-myc.
a,b, Heatmaps showing normalized expression levels of differentially expressed genes from selected enriched GO terms. Genes shown are differentially regulated by Casp1−⁄− Casp11−⁄− in both Eμ-myc and WT backgrounds. a, Genes in response to TGF-β, positive regulation of vasculature development, and cell-cell signaling by Wnt are shown. b, Genes in response to cellular respiration, mitochondrial respiratory chain complex assembly, mitochondrial organization, mitochondrial ATP synthesis coupled electron transport, and protein targeting to mitochondria are shown. Color scale indicates row z score of normalized read counts. c, Gene Ontology (GO) biological process terms most significantly enriched among genes differentially expressed due to Eμ-myc in both Casp1−⁄− Casp11−⁄− and wild type backgrounds. Common genes indicated in purple in Venn diagrams in Fig. 3c). Enrichment p values were corrected using the method of Benjamini and Hochberg.
Extended Data Fig. 6 The stromal inflammasome controls HSC surface TNFR1/II and MIP receptors via Ras.
a, Cytokine Array (ab193659) map for the Membranes C3 (top) and C4 (bottom) showing the identities of the 96 cytokine targets probed for per membrane. b, Representative cytokine array blots of WT (left) vs Casp1−⁄− Casp11−⁄− (right) bulk bone marrow after 6 h incubation including array controls of Membrane C4. c, Quantification of the top differentially expressed cytokines from the cytokine array blots of WT vs Casp1−⁄− Casp11−⁄− bulk bone marrow after 6 h incubation. [below 10000 units normalized fluorescence]; n = 3 biological replicates per genotype; ns, p > 0.05, p* ≤ 0.05, p*** ≤ 0.001, p**** ≤ 0.0001; 2 WAY Anova with ordinary 2 WAY Anova and SIdak’s multiple comparisons posttest comparing every genotype column mean by cytokine row mean. d, Representative histograms of stroma (CD45–Sca-1–c-Kit–Lin–) expression of indicated receptors from 6 h incubation of bulk bone marrow of adult male WT, Casp1−⁄− Casp11−⁄− Eµ-myc Casp1−⁄− Casp11−⁄− mice and WT bone marrow treated with pan caspase inhibitor QVD-OPH for 2 h. All data are presented as mean values +/− SD.
Extended Data Fig. 7 Gene expression profiles of inflammasome components in the bone marrow stroma.
a. UMAP visualizations of CD45- (Ptprc-) osteoblast, perivascular, and vascular bone marrow niche cells (n = 19,418) taken from Tikhonova A.N. et al.38. Top: Labeled and color-coded according to algorithmically determined clusters (Seurat clusters). Bottom: Labeled and color-coded according to bone marrow niche (VECAD = vascular, LERP = perivascular, COL1A1 = osteoblast), as determined by the expression of niche marker genes Cdh5 (VECAD), Lepr, and Col1a1. b. UMAP visualization of bone marrow niche cells colored according to the normalized expression of niche marker genes (Cdh5 = vascular, Lepr = perivascular, Col1a1 = osteoblast). c. Violin plots of normalized expression of niche marker genes across all clusters of CD45- (Ptprc-) bone marrow niche cells. d. Table shows our manual cluster annotations of the original Seurat clusters generated from the scRNA-sequencing dataset by Tikhonova A.N. et al.38. Bone marrow endothelial cells (BMEC), mesenchymal stromal cell (MSC), leptin-receptor (LEPR), and osteoblasts. Sub-markers used were based on Tikhonova A.N. et al.1: BMEC: V1 Ly6ahigh and V2 Stab2high and indicated as LEPR+ when applicable for Seurat clusters 6 and 15. LepR MSC: sub-marked as O1, O2, O3 based on low co-expression of Col1a1 or V1, V2 based on low co-expression of VEcad. Osteoblasts: O1 Col16a1highTnnhigh, O2 Fbn1highIgf1high, O3 BglaphighCar3high, low for osteoblast maturation marker (Bglaplo) or when co-expressing Lepr (Seurat clusters 11 and 14). e. Dot plot of the expression of genes of interest (columns) in annotated CD45– (Ptprc–) bone marrow niche cell clusters (rows). Dot size indicates the percent of cells within a cluster that have non-zero expression of the corresponding gene. Dot color corresponds to scaled average normalized expression of a gene within a cluster.
Extended Data Fig. 8 Relative expression of inflammasome genes in bone marrow stromal cells versus bone marrow macrophages.
Average normalized expression values of genes of interest in select cell populations/clusters, including CD45– (Ptprc–) bone marrow niche cells and bone marrow derived macrophages. Relative gene expression in manually annotated Seurat clusters a, VE-Cad+ bone marrow endothelial cells (BMEC), b, LERP+ LeprMSC (Leptin receptor+ mesenchymal stromal cells), and c, COL1A1+ osteoblasts. Manual cluster annotations were conducted as outlined in Extended Data Fig. 7d.
Extended Data Fig. 9 Ras target gene expression levels in WT HSCs isolated from different inflammasome component deficient stroma bearing mice.
Quantitative real time PCR showing the fold change expression of Ras signaling associated, and cancer related genes relative to WT (normalized to B2M at n = 1) of enriched wild type donor CD45.1+ CD45.2– bone marrow HSC (Lin–Sca-1+c-Kit+CD48–) isolated from chimeric WT CD45.2 recipient vs indicated CD45.2+ recipient WT, Nlrp3−⁄−, Pycard−⁄−, Casp1−⁄−, Gsdmd−⁄−, Casp11−⁄−, and Casp1−⁄− Casp11−⁄− mice. n = 3 biological replicates per genotype; p* ≤ 0.05, p** ≤ 0.01; 2 WAY Anova. Only p values ≤ 0.05 are shown. All data are presented as mean values +/− SD.
Extended Data Fig. 10 Proposed model – The stromal inflammasome maintains tissue equilibrium by restraining Ras activity to impede premalignant-to-malignant transition.
In the absence of inflammasome effectors caspases 1 and 11 in the bone marrow stroma, HSCs exhibited a distinct Ras-transduction signature associated with increased activity in the Ras pathway and an elevated expression of transcripts associated with cancer. Additionally, there was an increased presence of receptors for cytokines, chemokines, and growth-factors on HSCs. These findings suggest a model whereby inflammasome deficiency within the stroma creates a state of heightened Ras-dependent mitogenic processes, which cooperate with Myc upon its deregulation to accelerate the rate of malignant transformation and B cell lymphoma development. Created using BioRender.com and exported under a paid subscription to BioRender.
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Data 1
Genes enriched in sorted LSK HSCs compared with other hematopoietic cell types. Excel file shows 322 genes whose expression is enriched specifically in our sorted LSK HSCs compared with all other hematopoietic cell types in the ImmGen database33.
Supplementary Data 2
Lists of genes in GO pathways associated with HSC caspase-1/11 deficiency or Eμ-myc transgene expression. Excel file shows three tabs: Extended clusters, Caspase effect and Eμ-myc effect on the differential expression of genes found from RNA-seq on isolated LSK HSCs from premalignant Eμ-myc and Eμ-myc Casp1−⁄− Casp11−⁄− and their nontransgenic counterpart WT and Casp1−⁄− Casp11−⁄− mice (nontransgenic background).
Source data
Source Data Fig. 1
Statistical source data for the graphs in Fig. 1d–f, h–j and l.
Source Data Fig. 2
Statistical source data for the graphs in Fig. 2a and b.
Source Data Fig. 3
Statistical source data for the graphs in Fig. 3f–k.
Source Data Fig. 4
Statistical source data for the graphs in Fig. 4g and h.
Source Data Fig. 5
Statistical source data for the graphs in Fig. 5c, d, f, g and i.
Source Data Fig. 6
Statistical source data for the graphs in Fig. 6b, d and f–h.
Source Data Fig. 7
Statistical source data for the graphs in Fig. 7c, d, f, h and j.
Source Data Fig. 8
Statistical source data for the graphs in Fig. 8b, d,f, g and i.
Source Data Extended Data Fig. 1
Statistical source data for the graphs in Extended Data Fig. 1c, g and h.
Source Data Extended Data Fig. 2
Statistical source data for the graphs in Extended Data Fig. 2f and g.
Source Data Extended Data Fig. 3
Statistical source data for the graphs in Extended Data Fig. 3g and h.
Source Data Extended Data Fig. 6
Statistical source data for the graphs in Extended Data Fig. 6c.
Source Data Extended Data Fig. 9
Statistical source data for the graphs in Extended Data Fig. 9.
Source Data Fig. 6b and Extended Data Fig. 6b
Cytokine array blots in Fig. 6b and Extended Data Fig 6b.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kent, A., Yee Mon, K.J., Hutchins, Z. et al. A stromal inflammasome Ras safeguard against Myc-driven lymphomagenesis. Nat Immunol 26, 53–67 (2025). https://doi.org/10.1038/s41590-024-02028-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-024-02028-z