Abstract
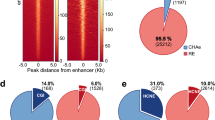
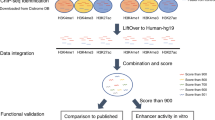
A major yet unresolved quest in decoding the human genome is the identification of the regulatory sequences that control the spatial and temporal expression of genes. Distant-acting transcriptional enhancers are particularly challenging to uncover because they are scattered among the vast non-coding portion of the genome. Evolutionary sequence constraint can facilitate the discovery of enhancers, but fails to predict when and where they are active in vivo. Here we present the results of chromatin immunoprecipitation with the enhancer-associated protein p300 followed by massively parallel sequencing, and map several thousand in vivo binding sites of p300 in mouse embryonic forebrain, midbrain and limb tissue. We tested 86 of these sequences in a transgenic mouse assay, which in nearly all cases demonstrated reproducible enhancer activity in the tissues that were predicted by p300 binding. Our results indicate that in vivo mapping of p300 binding is a highly accurate means for identifying enhancers and their associated activities, and suggest that such data sets will be useful to study the role of tissue-specific enhancers in human biology and disease on a genome-wide scale.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001)
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997)
Okazaki, Y. et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573 (2002)
Marshall, H. et al. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1 . Nature 370, 567–571 (1994)
Nobrega, M. A., Ovcharenko, I., Afzal, V. & Rubin, E. M. Scanning human gene deserts for long-range enhancers. Science 302, 413 (2003)
de la Calle-Mustienes, E. et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 15, 1061–1072 (2005)
Woolfe, A. et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 (2005)
Prabhakar, S. et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 16, 855–863 (2006)
Pennacchio, L. A. et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 (2006)
Visel, A. et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nature Genet. 40, 158–160 (2008)
Holland, L. Z. et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100–1111 (2008)
ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007)
Margulies, E. H. et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 17, 760–774 (2007)
Cooper, G. M. & Brown, C. D. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 18, 201–205 (2008)
McGaughey, D. M. et al. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 18, 252–260 (2008)
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods 4, 651–657 (2007)
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007)
Robertson, A. G. et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 18, 1906–1917 (2008)
Cuddapah, S. et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 (2009)
Wederell, E. D. et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 36, 4549–4564 (2008)
Valouev, A. et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nature Methods 5, 829–834 (2008)
Eckner, R. et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8, 869–884 (1994)
Eckner, R., Yao, T. P., Oldread, E. & Livingston, D. M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10, 2478–2490 (1996)
Yao, T. P. et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93, 361–372 (1998)
Merika, M., Williams, A. J., Chen, G., Collins, T. & Thanos, D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1, 277–287 (1998)
Maston, G. A., Evans, S. K. & Green, M. R. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7, 29–59 (2006)
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007)
Xi, H. et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 3, e136 (2007)
Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002)
Kothary, R. et al. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature 335, 435–437 (1988)
Visel, A., Minovitsky, S., Dubchak, I. & Pennacchio, L. A. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007)
Cheng, Y. et al. Transcriptional enhancement by GATA1-occupied DNA segments is strongly associated with evolutionary constraint on the binding site motif. Genome Res. 18, 1896–1905 (2008)
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
Hallikas, O. et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124, 47–59 (2006)
Pennacchio, L. A., Loots, G. G., Nobrega, M. A. & Ovcharenko, I. Predicting tissue-specific enhancers in the human genome. Genome Res. 17, 201–211 (2007)
Kim, T. H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005)
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008)
Schones, D. E. & Zhao, K. Genome-wide approaches to studying chromatin modifications. Nature Rev. Genet. 9, 179–191 (2008)
Barrera, L. O. et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 18, 46–59 (2008)
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008)
Kwok, R. P. et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370, 223–226 (1994)
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996)
Agalioti, T. et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103, 667–678 (2000)
Ge, K. et al. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417, 563–567 (2002)
Li, Z. et al. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl Acad. Sci. USA 100, 8164–8169 (2003)
Blow, M. J. et al. Identification of the source of ancient remains through genomic sequencing. Genome Res. 18, 1347–1353 (2008)
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002)
Krawchuk, D. & Kania, A. Identification of genes controlled by LMX1B in the developing mouse limb bud. Dev. Dyn. 237, 1183–1192 (2008)
Karolchik, D. et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 36, D773–D779 (2008)
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007)
Giardine, B. et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005)
Mikkelsen, T. S. et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177 (2007)
Acknowledgements
We wish to thank R. Hosseini and S. Phouanenavong for technical support, and J. Rubenstein, J. Long, J. Choi and Y. Zhu for help with microarray experiments. This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program and by the University of California, Lawrence Berkeley National Laboratory under contract no. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under contract no. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract no. DE-AC02-06NA25396. L.A.P. and E.M.R. were supported by the Berkeley-PGA, under the Programs for Genomic Applications, funded by National Heart, Lung, & Blood Institute, and L.A.P. by the National Human Genome Research Institute. A.V. was supported by an American Heart Association postdoctoral fellowship. B.R. was supported by grants from the National Human Genome Research Institute and the Ludwig Institute for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6 with Legends and Supplementary Tables 1, 6 and 7. (PDF 1271 kb)
Supplementary Tables
This file contains Supplementary Tables 2-4 which show p300 peaks in forebrain, midbrain, and limb (XLS 472 kb)
Supplementary Tables
This file contains Supplementary Table 5 which shows results of in vivo enhancer assays in transgenic mice (XLS 85 kb)
Supplementary Tables
This file contains Supplementary Tables 8-11 which show forebrain and limb over- and underexpressed genes (XLS 772 kb)
Rights and permissions
About this article
Cite this article
Visel, A., Blow, M., Li, Z. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009). https://doi.org/10.1038/nature07730
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07730
This article is cited by
-
Tissue-specific RNA Polymerase II promoter-proximal pause release and burst kinetics in a Drosophila embryonic patterning network
Genome Biology (2024)
-
Reporter gene assays and chromatin-level assays define substantially non-overlapping sets of enhancer sequences
BMC Genomics (2023)
-
SOX9 is a key component of RUNX2-regulated transcriptional circuitry in osteosarcoma
Cell & Bioscience (2023)
-
Superenhancers as master gene regulators and novel therapeutic targets in brain tumors
Experimental & Molecular Medicine (2023)
-
iEnhancer-DCLA: using the original sequence to identify enhancers and their strength based on a deep learning framework
BMC Bioinformatics (2022)