Abstract
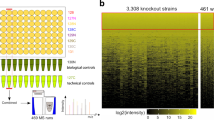
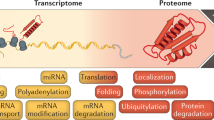
The availability of complete genomic sequences and technologies that allow comprehensive analysis of global expression profiles of messenger RNA1,2,3 have greatly expanded our ability to monitor the internal state of a cell. Yet biological systems ultimately need to be explained in terms of the activity, regulation and modification of proteins—and the ubiquitous occurrence of post-transcriptional regulation makes mRNA an imperfect proxy for such information. To facilitate global protein analyses, we have created a Saccharomyces cerevisiae fusion library where each open reading frame is tagged with a high-affinity epitope and expressed from its natural chromosomal location. Through immunodetection of the common tag, we obtain a census of proteins expressed during log-phase growth and measurements of their absolute levels. We find that about 80% of the proteome is expressed during normal growth conditions, and, using additional sequence information, we systematically identify misannotated genes. The abundance of proteins ranges from fewer than 50 to more than 106 molecules per cell. Many of these molecules, including essential proteins and most transcription factors, are present at levels that are not readily detectable by other proteomic techniques nor predictable by mRNA levels or codon bias measurements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lashkari, D. A. et al. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl Acad. Sci. USA 94, 13057–13062 (1997)
Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. A vision for the future of genomics research. Nature 422, 835–847 (2003)
Lockhart, D. J. et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnol. 14, 1675–1680 (1996)
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998)
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 17, 1030–1032 (1999)
Gavin, A. C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002)
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999)
Schwob, E., Bohm, T., Mendenhall, M. D. & Nasmyth, K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79, 233–244 (1994)
Grandin, N. & Reed, S. I. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 13, 2113–2125 (1993)
Huh, W.-K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003)
Harrison, P. M., Kumar, A., Lang, N., Snyder, M. & Gerstein, M. A question of size: The eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 30, 1083–1090 (2002)
Goffeau, A. Four years of post-genomic life with 6,000 yeast genes. FEBS Lett. 480, 37–41 (2000)
Das, S. et al. Biology's new Rosetta stone. Nature 385, 29–30 (1997)
Kowalczuk, M., Mackiewicz, P., Gierlik, A., Dudek, M. R. & Cebrat, S. Total number of coding open reading frames in the yeast genome. Yeast 15, 1031–1034 (1999)
Zhang, C. T. & Wang, J. Recognition of protein coding genes in the yeast genome at better than 95% accuracy based on the Z curve. Nucleic Acids Res. 28, 2804–2814 (2000)
Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 (2003)
Cliften, P. et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301, 71–76 (2003)
Ihmels, J. et al. Revealing modular organization in the yeast transcriptional network. Nature Genet. 31, 370–377 (2002)
Bergmann, S., Ihmels, J. & Barkai, N. Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E 67, 031902 (2003)
Gygi, S. P., Rochon, Y., Franza, B. R. & Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 (1999)
Futcher, B., Latter, G. I., Monardo, P., McLaughlin, C. S. & Garrels, J. I. A sampling of the yeast proteome. Mol. Cell. Biol. 19, 7357–7368 (1999)
Washburn, M. P. et al. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 100, 3107–3112 (2003)
Washburn, M. P., Wolters, D. & Yates, J. R. III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnol. 19, 242–247 (2001)
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003)
Holstege, F. C. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998)
Wang, Y. et al. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA 99, 5860–5865 (2002)
Sharp, P. M. & Li, W. H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (1987)
Grantham, R., Gautier, C. & Gouy, M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 8, 1893–1912 (1980)
Spellman, P. T. et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297 (1998)
Acknowledgements
We thank A. Carroll and F. Sanchez for technical assistance; J. Falvo, L. Gerke, J. Newman and members of the Weissman and O'Shea laboratories for discussions; and N. Barkai for providing data before publication. This work was supported by the Howard Hughes Medical Institute and the David and Lucile Packard Foundation. S.G. is a recipient of the Ruth L. Kirschstein National Research Service Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Ghaemmaghami, S., Huh, WK., Bower, K. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). https://doi.org/10.1038/nature02046
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02046