Abstract
Emerging data indicate that SARS-CoV-2-specific CD8+ T cells targeting different viral proteins are detectable in up to 70% of convalescent individuals1,2,3,4,5. However, very little information is currently available about the abundance, phenotype, functional capacity and fate of pre-existing and induced SARS-CoV-2-specific CD8+ T cell responses during the natural course of SARS-CoV-2 infection. Here, we define a set of optimal and dominant SARS-CoV-2-specific CD8+ T cell epitopes. We also perform a high-resolution ex vivo analysis of pre-existing and induced SARS-CoV-2-specific CD8+ T cells, applying peptide-loaded major histocompatibility complex class I (pMHCI) tetramer technology. We observe rapid induction, prolonged contraction and emergence of heterogeneous and functionally competent cross-reactive and induced memory CD8+ T cell responses in cross-sectionally analyzed individuals with mild disease following SARS-CoV-2 infection and three individuals longitudinally assessed for their T cells pre- and post-SARS-CoV-2 infection. SARS-CoV-2-specific memory CD8+ T cells exhibited functional characteristics comparable to influenza-specific CD8+ T cells and were detectable in SARS-CoV-2 convalescent individuals who were seronegative for anti-SARS-CoV-2 antibodies targeting spike (S) and nucleoprotein (N). These results define cross-reactive and induced SARS-CoV-2-specific CD8+ T cell responses as potentially important determinants of immune protection in mild SARS-CoV-2 infection.
Similar content being viewed by others
Main
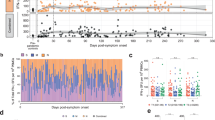
We selected five in silico predicted, SARS-CoV-2-derived epitope candidate peptides for each of the following human leukocyte antigen (HLA) class I alleles that are common in most populations worldwide (Extended Data Fig. 1): A*01:01, A*02:01, A*03:01, A*11:01 and A*24:02, as well as B*07:02, B*08:01, B*15:01, and B*40:01 and eight epitope candidate peptides for B*44:02/03 (Supplementary Table 1). In addition, we included all 13 previously described SARS-CoV-1-specific CD8+ T cell epitopes that display 100% homology in SARS-CoV-26 (Supplementary Table 1). We tested these 66 epitope peptides in 26 individuals (of white ancestry) with convalescent mild SARS-CoV-2 infection (Supplementary Table 2) in HLA-matched peptide-specific cell cultures. Importantly, we could detect SARS-CoV-2-specific CD8+ T cell responses in 23 out of 26 (88.5%) individuals, targeting a median of four epitopes (range 1–12; Fig. 1a). This exceeds the previously reported high detection rate of T cell responses in up to 70% of convalescent individuals in different cohorts using peptide pools for T cell stimulation1,2,3,4,5, probably reflecting our non-competitive approach. Identified CD8+ T cell epitopes were restricted by both HLA types, HLA-A and HLA-B (Fig. 1b). Thirty-three of 53 (62.3%) SARS-CoV-2-specific epitope candidates predicted in our study could be confirmed (Supplementary Table 1, depicted in bold). The strongest responses were observed for epitopes A*01/ORF3a207–215, A*02/ORF3a139–147 and B*07/N105–113, with medians of 8.3, 8.4 and 62.6% of CD8+ T cells producing interferon (IFN)-γ after peptide-specific culture, respectively (Fig. 1c,d). Taking the protein length into account, we observed an over-representation of N- and ORF3a-specific CD8+ T cell responses (Fig. 1e). Despite this, the absolute majority of detectable responses (57/110, 51.8%) targeted ORF1ab (Fig. 1e). This finding highlights the broad recognition of SARS-CoV-2 by virus-specific CD8+ T cells shown in earlier work1. Our approach to define optimal CD8+ T cell epitopes has the limitation that it does not completely cover the entire viral genome, in contrast to studies that have used overlapping peptides1,5,7, and therefore does not allow general assessment of immunodominance. Of note, we were able to detect SARS-CoV-2-specific CD8+ T cell responses in eight convalescent individuals that were seronegative for anti-SARS-CoV-2 S and N immunoglobulin-G (IgG) (Fig. 1f). To determine whether the identified SARS-CoV-2-specific CD8+ T cell epitopes are unique to SARS-CoV-2-exposed individuals, we tested historic blood samples (obtained before August 2019) from a cohort of 25 healthy volunteers with gender, age and HLA type characteristics comparable to our SARS-CoV-2 cohort (Supplementary Table 2). We observed very low virus-specific IFN-γ+ and tumor necrosis factor-positive (TNF+) CD8+ T cell responses in six individuals (five individuals, single response; one individual, five responses) (Fig. 1g, Supplementary Table 1 and Extended Data Fig. 2a) and TNF without IFN-γ responses in an additional four individuals (Extended Data Fig. 2a,c). The only epitope that was targeted by IFN-γ-secreting CD8+ T cells in more than one SARS-CoV-2-naive individual was epitope B*07/N105–113 (Extended Data Fig. 2a). This is the SARS-CoV-2-specific epitope in our study with the highest conservation between SARS-CoV-2 and ‘common cold’ coronaviruses (Extended Data Fig. 2b and Supplementary Table 3).
a, Pie chart illustrating the number of epitopes recognized per tested individual. b, Confirmed epitopes and total positive responses, depicted according to their HLA restriction. c, Representative dot plots showing peptide-loaded major histocompatibility complex class I (pMHCI) tetramer stainings and IFN-γ production of A*02/ORF3a139–147- and B*07/N105–113-specific CD8+ T cells after 14 days of in vitro expansion. Numbers refer to the respective percentage of pMHCI-tetramer+ and IFN-γ+ cells among CD8+ T cells. d, Percentage of convalescent SARS-CoV-2 individuals with a positive response toward HLA-A- and HLA-B-restricted SARS-CoV-2 peptides and the strength of individual responses as percent IFN-γ+ of CD8+ T cells. e, Confirmed epitopes and total positive responses depicted according to their location within the SARS-CoV-2 genome. f, CD8+ T cell responses in anti-SARS-CoV-2 S IgG (EUROIMMUN test, detection limit: 1.2 a.u. ml−1) and anti-SARS-CoV-2 N IgG (Mikrogen test, detection limit: 24 a.u. ml−1) seronegative convalescent individuals as percentage of responses out of all peptides tested matching the patient’s HLA alleles. Days post symptom onset (d.p.s.) are indicated. g, Exemplary dot plots showing pMHCI-tetramer staining and IFN-γ production of HLA-B*07/N105–113-specific CD8+ T cells from a historic control after 14 days in vitro expansion. Numbers refer to the respective percentage of pMHCI-tetramer+ and IFN-γ+ cells among CD8+ T cells. Bar charts show the median with interquartile range (IQR). n = 26 convalescent individuals. NA, not applicable (asymptomatic).
Next, we analyzed ex vivo SARS-CoV-2-specific CD8+ T cells targeting six of the above-identified optimal and dominant epitopes in comparison to influenza (FLU)-specific CD8+ T cells by using a set of peptide-loaded major histocompatibility complex class I (pMHCI) tetramers in a cohort of 20 convalescent individuals following a mild course of infection. Because the ex vivo frequencies of SARS-CoV-2-specific CD8+ T cells turned out to be low (Extended Data Fig. 3a), we performed pMHCI tetramer-based enrichment to increase the detection rate and to allow subsequent in-depth phenotypic analysis (Fig. 2a). After enrichment, we could detect SARS-CoV-2-specific CD8+ T cells in nearly all tested convalescent individuals (Fig. 2b and Extended Data Fig. 3b). SARS-CoV-2-specific CD8+ T cell frequencies were not associated with the time point of analysis during convalescence in relation to onset of symptoms (Fig. 2b and Extended Data Fig. 3c), indicating that the analyzed CD8+ T cell responses were within the late contraction/early memory phase. The calculated ex vivo frequencies of SARS-CoV-2-specific CD8+ T cells targeting A*01/ORF3a207–215, A*01/ORF1ab4163–4172, A*02/ORF3a139–147, B*44:03/N322–330 and B*44:03/ORF1ab3946–3954 were similar (Fig. 2b). CD8+ T cells targeting B*07/N105–113 were present in slightly higher frequencies compared to other SARS-CoV-2-specific CD8+ T cell populations, reaching the levels of A*02/Flu-M158–66-specific CD8+ T cells (Fig. 2b). SARS-CoV-2-specific CD8+ T cell frequencies were generally lower compared to cytomegalovirus (CMV) A*02/pp65496–503-specific and Epstein–Barr virus (EBV) A*02/ BMLF1280–288-specific CD8+ T cells in the context of latent viral infections associated with ‘inflationary memory’ CD8+ T cell responses8, but similar to the frequencies of virus-specific CD8+ T cells (targeting hepatitis B virus (HBV) A*02/pol455–463, HBV A*02/core18–27 and hepatitis C virus (HCV) A*02/NS31073–1081 and HCV A*02/NS31406–1415) in acutely resolved HBV and HCV infections (Extended Data Fig. 3d). SARS-CoV-2-specific CD8+ T cell populations in convalescent individuals were composed of naive (Tnaive), central memory (TCM), effector memory 1 (TEM1), effector memory 2 (TEM2), effector memory 3 (TEM3) and terminally differentiated effector memory expressing RA (TEMRA) T cell subsets, irrespective of the targeted epitope (Extended Data Fig. 3e,f). The presence of a minor Tnaive subset fraction (median, 3.9%) among all tested SARS-CoV-2-specific CD8+ T cells supports the notion that most of these cells have been efficiently primed during the infection. HLA-A- compared to HLA-B-restricted SARS-CoV-2-specific CD8+ T cells showed a shift toward the early differentiated TCM and TEM1 subset (Extended Data Fig. 3f). Similar results were obtained by applying the CX3CR1-based definition of memory T cell subsets (Extended Data Fig. 3g). In addition, t-distributed stochastic neighbor embedding (t-SNE) and multidimensional scaling (MDS) of all analyzed SARS-CoV-2-specific CD8+ T cells from the tested convalescent individuals (Fig. 2c) separated these cells more clearly according to their HLA restriction compared to the targeted viral protein. HLA-A-restricted SARS-CoV-2-specific CD8+ T cells were characterized by a cluster of markers including CD38, PD-1 and TOX, which are associated with antigen recognition and less differentiated cells expressed CD28 and T cell–specific transcription factor-1 (TCF-1) (Fig. 2d). By contrast, HLA-B-restricted SARS-CoV-2-specific CD8+ T cells clustered based on CD45RA, CD57, KLRG-1, CD25, CX3CR1 and high T-bet expression probably reflect a more terminally differentiated effector cell state (Fig. 2d and Extended Data Fig. 4a). Of note, FLU-A*02/M158–66-specific CD8+ T cells showed differences compared to HLA-A and HLA-B-restricted SARS-CoV-2-specific CD8+ T cells (Fig. 2c,d). In particular, FLU A*02/M158–66-specific CD8+ T cells expressed higher levels of CD127 and B cell lymphoma-2 (BCL-2), but TCF-1 and Forkhead box protein O1 (FOXO1) were similarly expressed (Fig. 2e and Extended Data Fig. 4b). The reduced BCL-2 expression of SARS-CoV-2-specific CD8+ T cells was most prominent among the early differentiated TCM and TEM1 subsets that have the highest BCL-2 expression among memory T cell subsets in general (Extended Data Fig. 4c). BCL-2 expression of SARS-CoV-2-specific CD8+ T cells correlated with time post-onset of symptoms (Fig. 2f). Thus, SARS-CoV-2-specific CD8+ T cells were probably not within a resting state but within the dynamic process of establishing a long-lasting memory compartment.
a, Epitope-specific CD8+ T cells ex vivo after pMHCI tetramer-based enrichment (10 to 20 × 106 peripheral blood mononuclear cells, PBMCs). Calculated ex vivo frequencies are depicted. b, Detection rates (left) and frequencies (middle) of epitope-specific T cells and their correlation with d.p.s. (right). c, t-SNE representation of fluorescence-activated cell sorting (FACS) data comparing T cells by HLA restriction (left) and targeted viral proteins (middle). MDS analysis results are also shown comparing HLA-A- and HLA-B-restricted epitopes (right). d, Expression levels (blue, low; red, high) of the indicated markers plotted on t-SNE plots. e, median fluorescence intensity (MFI) of indicated markers on virus-specific T cells normalized to MFI of naive T cells (nMFI). f, Correlation of BCL-2 expression with d.p.s. Bar charts show median with IQR. n = 6 (A*01/ORF3a207–215, A*01/ORF1ab4163–4172), n = 12 (A*02/ORF3a139–147), n = 7 (B*07/N105–113), n = 3 (B*44/N322–330, B*44/ORF1ab3946–3954), n = 5 (A*02/Flu-M158–66). Kruskal–Wallis rank-sum tests including Dunn’s multiple comparisons and Spearman correlation were performed.
To better determine the dynamics of the CD8+ T cell response in SARS-CoV-2 infection, we longitudinally analyzed SARS-CoV-2-specific CD8+ T cells in single patients. We had the unique opportunity to longitudinally follow the SARS-CoV-2-specific CD8+ T cell response before, during and after SARS-CoV-2 infection in an HLA-B*44:03+ individual with a defined infection event and documented onset of symptoms (Fig. 3a). B*44:03/N322–330- and B*44:03/ORF1ab3946–3954-specific CD8+ T cells were clearly expanded as early as seven days post infection (day 1 (d1) after symptom onset; Fig. 3b and Extended Data Fig. 5a). Both T cell populations were not detectable prior to SARS-CoV-2 infection (Fig. 3b and Extended Data Fig. 5a). The kinetics of both T cell responses were similar and the contraction phase lasted at least 70 days, with SARS-CoV-2-specific CD8+ T cells still detectable (frequencies of ~1 × 10−5) 104 days post symptom onset. The serum anti-SARS-CoV-2 N IgG titer reached the detection limit from day 29 post-symptom onset onward and the anti-SARS-CoV-2 S IgG titer fell below the detection limit at 79 days post symptom onset (Fig. 3c), while virus-specific CD8+ T cells remained detectable after enrichment at these same time points and also at later follow-up time points. Next, we performed deep profiling of SARS-CoV-2-specific CD8+ T cells, assessing expression of T cell differentiation and activation markers, transcription factors, inhibitory receptors and pro-survival factors with flow and mass cytometry. Diffusion map embedding combining flow cytometry data of B*44:03/N322–330- and B*44:03/ORF1ab3946–3954-specific CD8+ T cells indicated a continuous relationship between all SARS-CoV-2-specific CD8+ T cells collected during and after infection. Cells from early time points after symptom onset and those from later time points localize at opposing ends in the diffusion map, reflecting a dynamic differentiation of the virus-specific CD8+ T cell response (Fig. 3d and Extended Data Fig. 5b–d). Based on the linearity of the differentiation program suggested by the diffusion map analysis, we performed single-cell trajectory detection using Wanderlust analysis9 of mass cytometry data to understand the differentiation trajectories in more detail (Extended Data Fig. 6a). This analysis showed that a small fraction of virus-specific T cells identified one week after symptom onset with a CD28+ TCF-1+ CD127+ CD45RA+ phenotype may represent the precursor population of the large pool of effector cells (Extended Data Fig. 6a). As indicated by these Wanderlust (Extended Data Fig. 6a) and diffusion map (Fig. 3d) analyses, phenotyping by accelerated refined community-partitioning (PARC)10 of mass cytometry data confirmed a significant shift of SARS-CoV-2-specific CD8+ T cells from an early effector state (characterized by CD38, CD39 or PD-1 together with Ki-67) toward a TEM differentiation program (CD45RA, CX3CR1, KLRG-1, CD57) with little involvement of TCM cells (Fig. 3e and Extended Data Fig. 6b–d). These changes were also apparent on non-pMHCI tetramer+ CD8+ T cells (Extended Data Fig. 6b,e), suggesting broad activation of virus-specific responses targeting other epitopes. This is supported by additional longitudinal analyses of virus-specific CD8+ T cell responses in an HLA-A*02+ individual (up to 45 days post symptom onset, Fig. 3f) and an HLA-A*01+ individual (up to 21 days post symptom onset, Fig. 3g) pre- and post-infection with SARS-CoV-2 also demonstrating vigorous activation of A*02/ORF3a139–147-, A*01/ORF3a207–215- and A*01/ORF1ab4163–4172-specific CD8+ T cells (Fig. 3f,g and Extended Data Fig. 7a). Similar to the B*44:03/N322–330- and B*44:03/ORF1ab3946–3954-specific CD8+ T cells of the first case, A*02/ORF3a139–147-, A*01/ORF3a207–215- and A*01/ORF1ab4163–4172-specific CD8+ T cells exhibited elevated expression of the activation markers CD69 and CD38 at early time points that decreased over time, while BCL-2, CD127 and CD57 expression increased (Extended Data Fig. 7b). Anti-SARS-CoV-2 S and N IgG were detectable at all analyzed time points during SARS-CoV-2 infection in the last two cases, but these individuals were only followed until day 45 or day 21 post symptom onset, respectively (Fig. 3f,g). Altogether, these data show that SARS-CoV-2-specific CD8+ T cells restricted by different HLA alleles and targeting different epitopes are rapidly activated and expand during SARS-CoV-2 infection.
Longitudinal analyses are shown of SARS-CoV-2-infected individual D1. a, Timeline showing bleed dates (red arrowheads: light red, early time points; dark red, late time points), symptoms (dark gray bar) and positive PCR testing depicted at d.p.s. The infection time point is indicated with six days prior to symptom onset. b, The calculated ex vivo frequencies of B*44/N322–330- and B*44/ORF1ab3946–3954-specific CD8+ T cells (10 to 20 × 106 PBMCs) are indicated versus d.p.s. together with a historic sample. The dashed line indicates the detection threshold. c, Anti-SARS-CoV-2 S IgG (black dots; EUROIMMUN test, detection limit: 1.2 a.u. ml−1) and anti-SARS-CoV-2 N IgG (gray squares; Mikrogen test, detection limit: 24 a.u. ml−1) are depicted versus d.p.s. The dashed line indicates the detection limit of both tests. d, Diffusion map showing flow cytometry data of B*44/N322–330- and B*44/ORF1ab3946–3954-specific CD8+ T cells in relation to d.p.s. Protein expression levels are plotted on the diffusion map. e, The dynamic expression profile of SARS-CoV-2-specific CD8+ T cells, visualized as a heatmap. Data were collected by mass cytometry. Heatmap coloring represents percent of virus-specific CD8+ T cells expressing a given marker (blue, low expression; red, high expression). f, Longitudinal analysis of SARS-CoV-2-infected individual D2. The timeline shows bleed dates, symptoms and positive PCR testing depicted at d.p.s. (left). The frequency of A*02/ORF3a139–147-specific T cells (middle) and anti-SARS-CoV-2 S IgG and anti-SARS-CoV-2 N IgG (right) are depicted at d.p.s. together with a historic sample. g, Longitudinal analysis of SARS-CoV-2-infected individual D3. The timeline shows bleed dates, symptoms and positive PCR testing depicted at d.p.s. (left). The frequency of A*01/ORF3a207–215- and A*01/ORF1ab4163–4172-specific T cells (middle) and anti-SARS-CoV-2 S IgG and anti-SARS-CoV-2 N IgG (right) are depicted versus d.p.s. together with a historic sample.
We then assessed the functional capacity of SARS-CoV-2-specific compared to FLU-specific memory CD8+ T cells in vitro (Fig. 4a). After two weeks of in vitro expansion, we detected comparable frequencies of SARS-CoV-2 B*07/N105–113- and FLU A*02/M158–66-specific CD8+ T cells that were higher compared to the other tested SARS-CoV-2-specific CD8+ T cells (Fig. 4b, left). However, when analyzing the expansion index, a measure taking the input number of virus-specific CD8+ T cells into account, we observed comparable in vitro expansion capacities of the analyzed SARS-CoV-2- and FLU-specific CD8+ T cells (Fig. 4b, right). We also analyzed cytokine production (IFN-γ and TNF) and degranulation (CD107a) in relation to the frequency of virus-specific CD8+ T cells after expansion to obtain an approximation for the effector functions of SARS-CoV-2-specific CD8+ T cells. The functional capacity of SARS-CoV-2-specific CD8+ T cells was similar to A*02/Flu-M158–66-specific CD8+ T cells, irrespective of the targeted epitope (Fig. 4c). In the above-described HLA-B*44:03+ individual (Fig. 3a), we did not detect major changes in the in vitro functional capacity of SARS-CoV-2- specific CD8+ T cell populations within a time span of more than 100 days post symptom onset (Extended Data Fig. 8a–c). Together, these findings suggest that SARS-CoV-2-specific CD8+ T cells differentiate into functionally competent bona fide memory T cells comparable to FLU-specific CD8+ T cells, which represent classical, fully functional memory T cells11.
a, Workflow illustrating peptide-specific in vitro expansion of CD8+ T cells. b, Percentage of virus-specific CD8+ T cells (left) and expansion index (right) of the respective epitope-specific CD8+ T cells after in vitro expansion. c, Dot plots showing SARS-CoV-2-specific CD8+ T cells, and IFN-γ-, TNF- and CD107a-producing CD8+ T cells after in vitro expansion (top). The percentages of IFN-γ-, TNF- and CD107a-producing CD8+ T cells in relation to the frequency of epitope-specific CD8+ T cells were determined (bottom). d, Left: Dot plot showing virus-specific CD8+ T cells ex vivo after B*07/N105–113 tetramer-based enrichment (with 10 to 20 × 106 PBMCs; left). Calculated ex vivo frequencies of virus-specific CD8+ T cells are shown. Right: Pie chart depicting the number of positive responses of patients tested (middle). Frequency of B*07/N105–113-specific CD8+ T cells in historic controls compared to convalescent SARS-CoV-2 (cSARS-CoV-2) individuals (right). e, Distribution of CD8+ T cell subsets (Tnaive, TCM, TEM1, TEM2, TEM3 and TEMRA) of B*07/N105–113-specific CD8+ T cells in historic controls compared to convalescent SARS-CoV-2 individuals. f, Left: Dot plot showing virus-specific CD8+ T cells after in vitro expansion. The percentage of pMHCI tetramer+ cells among CD8+ T cells is depicted. Middle: Frequency and expansion index of virus-specific CD8+ T cells. Right: Expression of IFN-γ and TNF relative to the frequency of epitope-specific CD8+ T cells in historic controls compared to SARS-CoV-2 convalescent individuals. Historic controls were selected for a history of ‘common cold’ coronavirus infection. The bar charts show the medians with IQR. In b,c, n = 6 (A*01/ORF3a207–215, A*01/ORF1ab4163–4172), n = 12 (A*02/ORF3a139–147), n = 7 (B*07/N105–113), n = 3 (B*44/N322–330, B*44/ORF1ab3946–3954), n = 5 (A*02/Flu-M158–66). In d–f, n = 7 (convalescent SARS-CoV-2), n = 5 (d,e) or n = 3 (f) (historic controls). Statistical significance was assessed by Kruskal–Wallis rank-sum testing including Dunn’s multiple comparisons test.
Finally, we evaluated whether SARS-CoV-2 B*07/N105–113-specific memory CD8+ T cell responses differ between SARS-CoV-2 convalescent individuals (collected between 17 and 100 days post symptom onset, Extended Data Fig. 3b) and common cold coronaviruses-exposed individuals. To do this, we analyzed SARS-CoV-2 B*07/N105–113-specific CD8+ T cells in historic blood samples of 10 healthy B*07:02+ individuals that had tested positive for anti-common cold coronavirus N IgG (Fig. 4d and Supplementary Table 2). After pMHCI-based enrichment, we detected B*07/N105–113-specific CD8+ T cells ex vivo (5 of 10), but at lower frequencies compared to SARS-CoV-2 convalescent individuals (Fig. 4d). The B*07/N105–113-specific CD8+ T cell populations analyzed in SARS-CoV-2 convalescent individuals are probably within the late contraction/early memory phase, because no correlation of the frequency with respect to the timing of analysis was detectable (Extended Data Fig. 8d). The CD45RA/CCR7/CD27-based T cell subset distribution revealed a slight shift toward the further differentiated TEM3 subset in SARS-CoV-2 convalescent individuals (Fig. 4e). This further differentiation together with the higher ex vivo frequencies of B*07/N105–113-specific CD8+ T cells may indicate heterologous stimulation. Still, the B*07/N105–113-specific CD8+ T cell frequency was low, especially when comparing to cross-reactive virus-specific CD8+ T cell responses in the context of other viral infections in humans12. We did not observe differences in expansion and cytokine production of B*07/N105 113-specific CD8+ T cell populations in SARS-CoV-2 convalescent individuals compared to historic healthy controls (Fig. 4f).
Altogether, our findings indicate that pre-existing and induced SARS-CoV-2-specific CD8+ T cells represent major determinants of immune protection on an individual as well as population level. Whether our observation that SARS-CoV-2-specific CD8+ T cells were detectable in individuals seronegative for anti-SARS-CoV-2 S or N IgG indicates a faster waning of the antibody response compared to the CD8+ T cell response in SARS-CoV-2 infection, as has been reported for SARS-CoV-1 infection13,14, needs to be investigated in further detail and in larger cohorts. This requires longitudinal high-resolution analysis of SARS-CoV-2-specific antibodies targeting different viral proteins and of SARS-CoV-2-specific T cells and B cells15,16,17. In this study, we have now established experimental tools for high-resolution ex vivo analyses of SARS-CoV-2-specific CD8+ T cells that will also help to answer the question about the pathogenic versus protective role of virus-specific CD8+ T cells in SARS-CoV-2 infection. In future work, it will be important to evaluate whether differences in pre-existing and induced SARS-CoV-2-specific CD8+ T cell responses are linked to different courses of infection.
Methods
Study cohort
A total of 26 convalescent individuals (following a mild course of SARS-CoV-2 infection) and 25 age- and sex-matched historic controls (collected before August 2019) of healthy individuals (including pre-infection samples of longitudinal cases) were recruited at the Freiburg University Medical Center, Germany. A mild course of infection was defined as clinical symptoms without signs of respiratory insufficiency. The donor characteristics are summarized in Supplementary Table 2. SARS-CoV-2 infection was confirmed by positive PCR testing from oropharyngeal swab and/or SARS-CoV-2 spike IgG positive antibody testing in the presence of typical symptoms. pMHCI tetramer-based magnetic bead enrichment of virus-specific CD8+ T cells was performed with samples from 18 SARS-CoV-2 convalescent individuals and 10 historic controls. HLA typing was performed by next-generation sequencing and is presented in Supplementary Table 2. Influenza (FLU A*02/M158–66)-specific CD8+ T cell characterization was performed in five SARS-CoV-2 convalescent individuals, CMV (A*02/pp65496–503)- and EBV (A*02/BMLF1280–288)-specific CD8+ T cells were obtained from 23 healthy individuals and HBV (A*02/pol455–463, A*02/core18–27)-specific and HCV (A*02/NS31073–1081, A*02/NS31406–1415)-specific CD8+ T cells were analyzed from donors with an acutely resolved HBV (n = 7) or HCV (n = 5) infection. Written informed consent was obtained from all participants and the study was conducted according to federal guidelines, local ethics committee regulations (Albert-Ludwigs-Universität, Freiburg, Germany; no. 322/20) and the Declaration of Helsinki (1975).
PBMC isolation
Venous blood samples were collected in ethylenediaminetetraacetic acid-anticoagulated tubes. PBMCs were isolated with lymphocyte separation medium density gradients (Pancoll separation medium, PAN Biotech) and resuspended in RPMI 1640 medium supplemented with 10% FCS, 1% penicillin/streptomycin and 1.5% HEPES buffer 1 mol l−1 (complete medium; all additives from Thermo Scientific) and stored at −80 °C until used.
Prediction of SARS-CoV-2-specific CD8+ T cell epitopes
The entire viral amino-acid sequence of SARS-CoV-2 (GenBank: MN908947.3) was analyzed for in silico peptide binding with ANN 4.0 on the Immune Epitope Database website18. The five best 8-, 9- or 10-mer peptides calculated for HLA alleles A*01:01, A*02:01, A*03:01, A*11:01, A*24:02, B*07:02, B*08:01, B*15:01, B*40:01 and B*44:02/03 were selected and synthesized for further analysis. Additionally, 13 epitopes that were predicted by Grifoni et al. with high sequence similarity to SARS-CoV-1 were included, as summarized in Supplementary Table 16.
Sequence alignment
Sequence homology analyses were performed in Geneious Prime 2020.0.3 (https://www.geneious.com/) using Clustal Omega 1.2.2 alignment with default settings19. Reference genomes of human coronaviruses were downloaded from NCBI databases 229E (NC_002645), HKU1 (NC_006577), NL63 (NC_005831), OC43 (NC_006213), MERS (NC_019843) and SARS-CoV-1 (NC_004718). Proteins of human coronaviruses were aligned according to their homology (amino-acid level) only if the protein of interest had a homolog in the respective coronavirus. Confirmed SARS-CoV-2 epitopes were then mapped to the corresponding protein alignment, as summarized in Supplementary Table 3.
Peptides and tetramers
Peptides were synthesized with an unmodified N terminus and an amidated C terminus with standard Fmoc chemistry and a purity of >70% (Genaxxon Bioscience). HLA class I easYmers (immunAware) were loaded with peptide according to the manufacturer’s instructions (A*01/ORF3a207–215, A*01/ORF1ab4163–4172, A*02/ORF3a139–147, B*07/N105–113) or ordered as peptide-loaded monomers (B*44:03/N322–330, B*44:03/ORF1ab3946–3954). SARS-CoV-2 peptide-loaded HLA class I tetramers were generated by conjugation of biotinylated peptide-loaded HLA class I easYmers with phycoerythrin (PE)-conjugated streptavidin (Agilent) according to the manufacturer’s instructions. Influenza-specific HLA-A*02/M158–66 (GILGFVFTL) tetramers, CMV-specific HLA-A*02/pp65496–503 tetramers, EBV-specific HLA-A*02/BMLF1280–288 tetramers, HBV-specific HLA-A*02/pol455–463 and HLA-A*02/core18–27 tetramers and HCV-specific HLA-A*02/NS31073–1081 and HLA-A*02/NS31406–1415 tetramers were generated as described previously20.
In vitro expansion of virus-specific CD8+ T cells and assessment of effector function
PBMCs (1−2 × 106) were stimulated with epitope-specific peptides (5 µM) and anti-CD28 mAb (0.5 µg ml−1, BD Biosciences) and expanded for 14 days in complete RPMI culture medium containing rIL2 (20 IU ml−1, Miltenyi Biotec). The expansion factor was calculated based on peptide-loaded HLA class I tetramer staining, as described previously21. Cytokine production and degranulation were assessed 5 h after re-stimulation with epitope-specific peptides as previously described21.
Magnetic bead-based enrichment of antigen-specific CD8 T cells
Enrichment of virus-specific CD8+ T cells was performed as described previously22. Briefly, 1 to 2 × 107 PBMCs (with an average of 16.5% CD8+ T cells) were labeled for 30 min with PE-coupled peptide-loaded HLA class I tetramers. Subsequent enrichment was performed with anti-phycoerythrin (PE) beads using magnetic-activated cell sorting (MACS) technology (Miltenyi Biotec) according to the manufacturer’s protocol. Enriched SARS-CoV-2-specific CD8+ T cells were used for multiparametric flow cytometry analysis. Frequencies of virus-specific CD8+ T cells were calculated as described previously22, with a detection limit of 2 × 10−6.
Multiparametric flow cytometry
The following antibodies were used for multiparametric flow cytometry: anti-CCR7-PE-CF594 (cat. no. 150503, dilution 1:50), anti-CCR7-BUV395 (cat. no. 3D12, dilution 1:50), anti-CCR7-BV421 (cat. no. 150503, dilution 1:33), anti-CD4-BV786 (cat. no. L200, dilution 1:200), anti-CD8-BUV496 (cat. no. SK1, dilution 1:100), anti-CD8-BUV510 (cat. no. SK1, dilution 1:100), anti-CD8-APC (cat. no. SK-1, dilution 1:200), anti-CD27-BV605 (cat. no. L128, dilution 1:200), anti-CD28-BV421 (cat. no. CD28.2, dilution 1:100), anti-CD28-BV711 (cat. no. CD28.2, dilution 1:100), anti-CD45RA-BV786 (cat. no. HI100, dilution 1:800), anti-CD45RA-BUV737 (cat. no. HI100, dilution 1:200), anti-CD69-BUV395 (cat. no. FN50, dilution 1:50), anti-CD107a-APC (cat. no. H4A3, dilution 1:100), anti-CD127-BV510 (cat. no. HIL-7R-M21, dilution 1:25), anti-EOMES-PerCP-eF710 (cat. no. WD1928, dilution 1:50), anti-IFN-γ-FITC (cat. no. 25723.11, dilution 1:8), anti-IL-21-PE (cat. no. 3A3-N2.1, dilution 1:25), anti-PD-1-BV786 (cat. no. EH12.1, dilution 1:33) and anti-TNF-PE-Cy7 (cat. no. Mab11, dilution 1:400) (all obtained from BD Biosciences); anti-BCL-2-BV421 (cat. no. 100, dilution 1:200), anti-CD25-BV650 (cat. no. BC96, dilution 1:33), anti-CD38-BV650 (cat. no. HB-7, dilution 1:400), anti-CD57-BV605 (cat. no. QA17A04, dilution 1:100), anti-CX3CR1-APC-eFluor660 (cat. no. 2A9-1, dilution 1:50), anti-CXCR3-PerCP-Cy5.5 (cat. no. G025H7, dilution 1:33), anti-IL-2-PerCP-Cy5.5 (cat. no. MQ1-17H12, dilution 1:100), anti-IL17A-BV605 (cat. no. BL168, dilution 1:100), anti-PD-1-PE-Cy7 (cat. no. EH12.2H7, dilution 1:200), anti-rabbit-PE-CF594 (cat. no. Poly4064, dilution 1:200) and anti-CD45RA-BV510 (cat. no. HI100, dilution 1:200) (all obtained from BioLegend); anti-FOXO-1-pure (cat. no. C29H4, dilution 1:33) and anti-TCF-1-AlexaFluor488 (cat. no. C63D9, dilution 1:100) (Cell Signaling); anti-CD14-APC-eFluor780 (cat. no. 61D3, dilution 1:400), anti-CD19-APC-eFluor780 (cat. no. HIB19, dilution 1:400), anti-CD27-FITC (cat. no. 0323, dilution 1:100), anti-KLRG-1-BV711 (cat. no. 13F12F2, dilution 1:50), anti-T-bet-PE-Cy7 (cat. no. 4B10, dilution 1:200) and anti-TOX-eFluor660 (cat. no. TRX10, dilution 1:100) (eBioscience). A fixable viability dye (eBioscience; APC-eFluor780 dilutions 1:200, 1:400) or ViaProbe (BD Biosciences; cat. no. 7-AAD, dilution 1:33) was used for live/dead discrimination. A FoxP3/transcription factor staining buffer set (eBioscience) and fixation/permeabilization solution kit (BD Biosciences) were used according to the manufacturers’ instructions to stain for intranuclear and cytoplasmic molecules, respectively. Fixation of cells in 2% paraformaldehyde (PFA, Sigma) was followed by subsequent analyses on FACSCanto II, LSRFortessa (BD Biosciences) or CytoFLEX (Beckman Coulter) systems. Data analyses were performed with FlowJo 10 (TreeStar).
Dimensionality reduction of multiparametric flow cytometry data
The visualization of multiparametric flow cytometry data was done with R version 4.0.2 using the Bioconductor (release (3.11)) CATALYST package23. The analyses were performed on gated virus-specific CD8+ T cells for two panels separately. Analysis of panel 1 (transcription factors) included the markers CD45RA, CCR7, CD27, CD28, BCL-2, TCF-1, CD69, CD38, PD-1, EOMES, T-bet and TOX. Analysis of panel 2 (surface markers) was performed on CCR7, CD45RA, CD27, CD28, CD25, CD127, CD57, KLRG-1, CXCR3, PD-1, CX3CR1 and FOXO-1. Downsampling of cells to the number of cells present in the sample with the fewest cells was performed before dimensionality reduction to facilitate the visualization of different samples. Marker intensities were transformed by arcsinh (inverse hyperbolic sine) with a cofactor of 150. Dimensionality reduction on the transformed data was achieved by t-SNE, MDS and diffusion map visualization.
Mass cytometry
Mass cytometry reagents were obtained from Fluidigm or generated by custom conjugation to isotope-loaded polymers using a MAXPAR X8 conjugation kit (Fluidigm). The mass cytometry antibodies used are shown in Supplementary Table 2. Mass cytometry tetramers were generated by tetramerization of pMHCI monomers with streptavidin conjugated to Eu151 using a Lightning link conjugation kit (Expedon) Sample barcoding was performed using anti-β2M barcodes, then cells were pooled and staining was performed as previously described24. Briefly, the single-cell suspension was pelleted, incubated with 20 μM Lanthanum-139 (Trace Sciences)-loaded maleimido-mono-amine-DOTA (Macrocyclics) in phosphate-buffered saline (PBS) for 10 min at room temperature (RT) for live/dead discrimination. Cells were washed in staining buffer and resuspended in staining buffer containing tetramers, incubated for 30 min at RT and washed twice. Cells were then resuspended in surface antibody cocktail, incubated for 30 min at RT, washed twice in staining buffer, pre-fixed with PFA (Electron Microscopy Sciences) 1.6%, washed, then fixed and permeabilized using FoxP3 staining buffer set (eBioscience) and stained intracellularly for 60 min at RT. Cells were further washed twice before fixation in 1.6% PFA solution containing 125 nM iridium intercalator overnight at 4 °C. Before data acquisition on a CyTOF Helios (Fluidigm), cells were washed twice in PBS and once in cell acquisition solution (CAS; Fluidigm). Mass cytometry data were analyzed after debarcoding and bead-based normalization. For analysis of mass cytometric data, samples were first gated on iridium intercalator positive, live, single CD45+ CD3+ CD8+ T cells using FlowJo (v10.6). CD8+ T cells were then exported for analysis in Omiq (Omiq). Virus-specific CD8+ T cells were identified by manual gating. A workflow including dimension reduction using optSNE25, PARC clustering analysis10 and Wanderlust trajectory analysis9 was implemented in Omiq. Clustering and dimension reduction analysis were performed based on CD45RA, CD45RO, CCR7, CD28, CD127, CD16, CD25, CD26, CD38, CD39, CD56, CD57, CD69, CD103, CD161, CCR6, CCR9, CXCR3, CXCR5, CXCR6, CX3CR1, CRTH2, TCF-1, TOX, TIGIT, T-bet, EOMES, KLRG-1 and PD-1. Further analysis and heatmap visualization was performed using R (v4.0) (https://www.r-project.org).
Serum IgG determination
SARS-CoV-2-specific antibodies were determined by the EUROIMMUN assay detecting anti-SARS-CoV-2 spike IgG (anti-SARS-CoV-2 S IgG; detection limit, 1.2 a.u. ml−1) and by the Mikrogen assay detecting anti-SARS-CoV-2 nucleocapside IgG (anti-SARS-CoV-2 N IgG; detection limit, 24 a.u. ml−1), as described in the product instructions. Infection with common cold coronaviruses 229E, NL63, OC43 and HKU1 was determined by the Mikrogen recomline assay detecting the anti-nucleocapside IgG of common cold coronaviruses 229E, NL63, OC43 and HKU1 (detection limit defined by the cutoff control provided by the supplier).
Statistics
Statistical analysis was performed with GraphPad Prism 8. Statistical significance was assessed by Kruskal–Wallis testing including Dunn’s multiple comparisons test and Spearman correlation (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Raw data for this study are provided as the supplementary dataset at https://github.com/sagar161286/SARSCoV2_specific_CD8_Tcells (flow cytometry data) and at https://flowrepository.org/experiments/3159 (mass cytometry data). All requests for additional supporting raw and analyzed data and materials will be reviewed by the corresponding authors to verify if the request is subject to any intellectual property or confidentiality obligations. Patient-related data not included in the paper were generated as part of clinical examination and may be subject to patient confidentiality. Any data and materials that can be shared will be released via a material transfer agreement. Reference viral sequences (SARS-CoV-2 (MN908947.3), 229E (NC_002645), HKU1 (NC_006577), NL63 (NC_005831), OC43 (NC_006213), MERS (NC_019843) and SARS-CoV-1 (NC_004718)) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). In silico peptide binding was analyzed with ANN 4.0 on the Immune Epitope Database website (https://www.iedb.org/). Source data are provided with this paper.
Code availability
R code to reproduce the analysis of multiparametric flow cytometry data is available at https://github.com/sagar161286/SARSCoV2_specific_CD8_Tcells. Detailed settings for dimension reduction, clustering and trajectory analysis of mass cytometry data is available via the platform Omiq.ai upon request. This request will be reviewed by the corresponding authors to verify if it is subject to any intellectual property or confidentiality obligations.
References
Grifoni, A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501 (2020).
Le Bert, N. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020).
Ni, L. et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52, 971–977 (2020).
Sekine, T. et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183, 158–168 (2020).
Peng, Y. et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 21, 1336–1345 (2020).
Grifoni, A. et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 27, 671–680 (2020).
Weiskopf, D. et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 5, eabd2071 (2020).
Klenerman, P. The (gradual) rise of memory inflation. Immunol. Rev. 283, 99–112 (2018).
Bendall, S. C. et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014).
Stassen, S. V. et al. PARC: ultrafast and accurate clustering of phenotypic data of millions of single cells. Bioinformatics 36, 2778–2786 (2020).
van de Sandt, C. E. et al. Human influenza A virus-specific CD8+ T-cell response is long-lived. J. Infect. Dis. 212, 81–85 (2015).
Urbani, S. et al. Heterologous T cell immunity in severe hepatitis C virus infection. J. Exp. Med. 201, 675–680 (2005).
Cao, W. C., Liu, W., Zhang, P. H., Zhang, F. & Richardus, J. H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 357, 1162–1163 (2007).
Ng, O. W. et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 34, 2008–2014 (2016).
Hachim, A. et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 21, 1293–1301 (2020).
Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020).
Juno, J. A. et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 26, 1428–1434 (2020).
Nielsen, M. et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 12, 1007–1017 (2003).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Price, D. A. et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 202, 1349–1361 (2005).
Wieland, D. et al. TCF1+ hepatitis C virus-specific CD8+ T cells are maintained after cessation of chronic antigen stimulation. Nat. Commun. 8, 15050 (2017).
Alanio, C., Lemaitre, F., Law, H. K., Hasan, M. & Albert, M. L. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood 115, 3718–3725 (2010).
Crowell, H., Zanotelli, V., Chevrier, S. & Robinson, M. CATALYST: Cytometry dATa anALYSis Tools. R package version 1.12.2 (2020); https://github.com/HelenaLC/CATALYST
Bengsch, B. et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity 48, 1029–1045 (2018).
Belkina, A. C. et al. Automated optimized parameters for t-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat. Commun. 10, 5415 (2019).
Acknowledgements
We thank all donors for participating in the current study and the FREEZE-Biobank Center for biobanking (Freiburg University Medical Center) and the Medical Faculty for support. The study was funded by the Federal Ministry of Education and Research (grant no. 01KI1722, to G.K., M.H., M.P., M.S. and R.T.) and by a COVID-19 research grant of the Ministry of Science, Research and Art, State of Baden-Wuerttemberg (to C.N.H. and B.B.). The presented work was also supported by CRC/TRR 179-Project 01 and CRC 1160-Project A02 (to R.T.), CRC/TRR 179-Project 02 and CRC 1160-Project A06 (to C.N.H.), CRC/TRR 179-Project 04 (to T.B.), CRC/TRR 179-Project 20 and CRC 1160-Project A02 (to M.H.), CRC/TRR 179-Project 21, CRC 1160-Project A03 and BE-5496/5-1 (to B.B.) of the German Research Foundation (DFG; TRR 179 project no. 272983813; CRC 1160 project no. 256073931). M.H. was supported by a Margarete von Wrangell fellowship (State of Baden-Wuerttemberg). D.B. and T.B. are supported by the Berta-Ottenstein Programme, Faculty of Medicine, University of Freiburg. H.E.M. was supported by DFG ME3644/5-1(project no. 324736178) and D.A.P. by a Wellcome Trust Senior Investigator Award (100326/Z/12/Z). The funding body had no role in the decision to write or submit the manuscript.
Author information
Authors and Affiliations
Contributions
I.S., J.K., V.O. and K.W. planned, performed and analyzed experiments with the help of S., F.D. and O.S. L.M.S. and S.K. performed and analyzed mass cytometry data with the assistance of M.S.L. A.D., H.L., B. Binder, D.B., S.R., C.F.W., A.N. and D.D. were responsible for donor recruitment. H.E.M. and A.R.S. provided barcoding reagents for mass cytometry analysis. S.L.-L. and D.A.P. provided influenza M158/A*02 tetramers. F.E. performed four-digit HLA typing by next-generation sequencing. M.P. and D.H. performed antibody testing. M.S. and G.K. provided virological expertise and contributed to data interpretation. B. Bengsch designed and supervised mass cytometry analysis. T.B., R.T., M.H. and C.N.-H. designed the study and contributed to experimental design and planning. I.S., J.K., V.O., R.T., M.H. and C.N.-H. interpreted data and wrote the manuscript. C.N.-H., M.H., R.T., B. Bengsch and T.B. are shared last authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 HLA distribution in different populations.
HLA-A and HLA-B distribution in different populations compared to the study population. Arrows indicate HLA alleles for which peptide epitope candidates were predicted and further analyzed.
Extended Data Fig. 2 Cross-reactivity of SARS-CoV-2-specific CD8+ T cells.
a, % of historic controls with IFN-γ response towards HLA-A- and HLA-B-restricted SARS-CoV-2 peptides and strength of individual responses as % IFN-γ+ and % TNF+ of CD8+ T cells. b, Heat map illustrating the degree of homology between confirmed SARS-CoV-2 epitopes and ‘common cold’ coronaviruses 229E, OC43, HKU1 and NL63 (bright green: no amino acid (aa) change, 100% homology; light green: 1 aa difference; yellow: 2 aa difference; orange: 3 aa difference; bright red: 4-10 aa difference, dark red: aa residue at HLA-binding anchor is different, resulting in an IC50 increase by a factor >10 calculated by ANN 4.0; black: no homolog sequence). c, Dot plot showing TNF production with and without SARS-CoV-2 peptide re-stimulation after 14-days in vitro expansion in a historic control. Bar charts show median with IQR. n=25 historic controls.
Extended Data Fig. 3 SARS-CoV-2-specific CD8+ memory T cell subsets.
a, Representative dot plots showing A*01/ORF3a207-215-, A*01/ORF1ab4163-4172-, A*02/ORF3a139-147-, B*07/N105-113-, B*44/N322-330- and B*44/ORF1ab3946-3954 -specific CD8+ T cells ex vivo after conventional pMHCI tetramer staining (1-2*106 PBMCs used). Numbers refer to the respective percentage of pMHCI-tetramer+ cells among CD8+ T cells. b, Cumulative depiction of the calculated ex vivo frequencies of virus-specific CD8+ T cells for each epitope analyzed in the corresponding donor. c, The day post symptom onset (dps) of the respective epitope-specific CD8+ T cells that were analyzed is depicted. d, The frequency of A*02/CMV-pp65495-503-, A*02/EBV-BMLF1280-288-, A*02/HBV-pol455-463-/core18-27- and A*02/HCV280-288-specific CD8+ T cells ex vivo after pMHCI tetramer-based enrichment (with 10-20*106 PBMCs) was determined. Dashed line indicates detection threshold. e, Gating strategy for CD8+ T-cell populations. f, Distribution of CD8+ T-cell subsets, naïve T cells (Tnaive), central memory T cells (TCM), effector memory T cells 1 (TEM1), effector memory T cells 2 (TEM2), effector memory T cells 3 (TEM3) and terminally differentiated effector memory cells re‐expressing CD45RA (TEMRA) among CD8+ T cells targeting the different epitopes. g, Exemplary histogram showing the gating of cell populations expressing no (neg) CX3CR1, intermediate (int) CX3CR1 and high (hi) CX3CR1 on CD8+ bulk (black) and non-naïve CD8+ T cells (grey) as well as SARS-CoV-2-specific CD8+ T cells (white). % of SARS-CoV-2-specific CD8+ T cells targeting the different epitopes expressing neg, int or hi levels of CX3CR1. f,g, Ex vivo analyses of virus-specific CD8+ T cells after pMHCI tetramer-based enrichment (10-20*106 PBMCs used for enrichment). Bar charts show the median with IQR. n=6 (A*01/ORF3a207-215, A*01/ORF1ab4163-417), n=12 (A*02/ORF3a139-147), n=7 (B*07/N105-113), n=3 (B*44/N322-330, B*44/ORF1ab3946-3954), n=5 (A*02/Flu-M158-66). Statistical significance was assessed by Kruskal-Wallis rank-sum testing including Dunn’s multiple comparisons test.
Extended Data Fig. 4 Phenotype of SARS-CoV-2-specific CD8+ T cells.
a, t-SNE representation of flow cytometric data, which were derived from 18 convalescent SARS-CoV-2 individuals after ex vivo pMHCI tetramer-based enrichment (10-20*106 PBMCs used for enrichment), comparing SARS-CoV-2-specific CD8+ T cells by their HLA restriction (left) and expression levels of CD127, CXCR3, FOXO-1, CX3CR1, CD57, KLRG-1 and CD25 plotted on the t-SNE plot. b, Exemplary histograms depicting the expression levels of CD127, BCL-2, TCF-1 and FOXO-1 on CD8+ bulk (black) and non-naïve CD8+ T cells (grey) as well as SARS-CoV-2-specific CD8+ T cells (white) ex vivo after pMHCI tetramer-based enrichment. c, BCL-2 expression of different memory cell populations on bulk CD8+ T cells (left) and of SARS-CoV-2- and Flu -specific CD8+ T cells ex vivo after pMHCI tetramer-based enrichment of the different CD8+ T-cell subsets. Bar charts show the median with IQR. n=6 (A*01/ORF3a207-215, A*01/ORF1ab4163-417), n=12 (A*02/ORF3a139-147), n=7 (B*07/N105-113), n=3 (B*44/N322-330, B*44/ORF1ab3946-3954), n=5 (A*02/Flu-M158-66). Statistical significance was assessed by Kruskal-Wallis or Friedman rank-sum C, testing including Dunn’s multiple comparisons test.
Extended Data Fig. 5 Longitudinal assessment of the SARS-CoV-2-specific CD8+ T-cell phenotype.
a, Dot plots show B*44/ORF1ab3946-3954-specific CD8+ T cells ex vivo after tetramer-based enrichment (10-20*106 PBMCs). Calculated ex vivo frequency of virus-specific CD8+ T cells is depicted. b, Histograms depicting the protein expression levels on CD8+ bulk (black), CD8+non-naïve (grey) and SARS-CoV-2-specific CD8+ T cells (white) and the expression of these markers on SARS-CoV-2-specific CD8+ T cells at the respective dps. c,d, Diffusion map of flow cytometric data, derived from longitudinal analysis from D1, showing B*44/N322-330- and B*44/ORF1ab3946-3954-specific T cells in relation to dps. Protein expression levels (color coded: blue = low expression; red = high expression) are plotted on the diffusion map. Dps are distinguished by a color gradient from light (early time points) to dark red (late time points) color (top left).
Extended Data Fig. 6 Deep longitudinal profiling of SARS-CoV-2-specific CD8+ T cells.
a, t-SNE map was calculated with all SARS-CoV-2-specific CD8+ T-cell data during (6 dps) and after (79 dps) infection (tet-t-SNE). Wanderlust trajectory analysis was performed on virus-specific CD8+ T cells. Wanderlust trajectory is indicated by heatmap colorization on the tet-t-SNE plot. Marker expression (color-coded: blue, low expression; red, high expression) is depicted according to wanderlust trajectory progression. b, The CD8 landscape in cSARS-CoV-2 was calculated using t-SNE on CD8+ T cells from before, during and after infection. Clustering was performed with PARC algorithm and clusters are indicated by the indicated color. c, SARS-CoV-2-specific CD8+ T cells are displayed on the t-SNE map. Frequency of virus-specific CD8+ T cells per CD8 cluster is illustrated using stacked bar chart. d, Hierarchically clustered heatmap of PARC cluster phenotypes – the indicated marker expression is shown per cluster as z-Score of median signal intensity per channel; blue, low expression; red, high expression. e, Frequency of each cluster of historic sample (circle), 6 dps (square) and 79 dps (triangle).
Extended Data Fig. 7 Additional longitudinal analysis of SARS-CoV-2-specific CD8+ T cells.
(a) Representative dot plots showing pMHCI tetramer stainings of A*02/ORF3a139-147- (for cSARS-CoV-2 individual D2) and A*01/ORF3a207-215-, A*01/ORF1ab4163-4172- specific CD8+ T cells (for cSARS-CoV-2 individual D3) ex vivo after tetramer-based enrichment (10-20*106 PBMCs used for enrichment) at different time points post symptom onset (dps). Numbers refer to the calculated ex vivo frequency of virus-specific CD8+ T cells. (b) Relative mean fluorescence intensity of proteins expressed on A*02/ORF3a139-147- (for cSARS-CoV-2 individual D2) and A*01/ORF3a207-215-, A*01/ORF1ab4163-4172- specific CD8+ T cells (for cSARS-CoV-2 individual D3) ex vivo after tetramer-based enrichment (10-20*106 PBMCs used for enrichment) at the respective days post symptom onset (dps).
Extended Data Fig. 8 Longitudinal SARS-CoV-2-specific CD8+ T-cell function.
a, Representative dot plots showing pMHCI tetramer stainings of HLA-B*44/N322-330 and HLA-B*44/ORF1ab3946-3954-specific CD8+ T cells (derived from D1) after 14 days in vitro expansion at different time points post symptom onset (dps). Numbers refer to the respective percentage of pMHCI tetramer+ cells out of CD8+ T cells b, Frequency and expansion index of virus-specific CD8+ T cells after 14 days in vitro expansion at dps. c, Expression of IFN-γ, TNF and CD107a/degranulation in percentage relative to the frequency of epitope-specific CD8+ T cells are depicted at dps. d, The calculated ex vivo frequency of B*07/N105-113 -specific CD8+ T cells (10-20*106 PBMCs used for enrichment) is indicated at dps. Statistical significance was assessed by Spearman correlation.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Schulien, I., Kemming, J., Oberhardt, V. et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med 27, 78–85 (2021). https://doi.org/10.1038/s41591-020-01143-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-020-01143-2
This article is cited by
-
The spike-specific TCRβ repertoire shows distinct features in unvaccinated or vaccinated patients with SARS-CoV-2 infection
Journal of Translational Medicine (2024)
-
Machine learning-enhanced immunopeptidomics applied to T-cell epitope discovery for COVID-19 vaccines
Nature Communications (2024)
-
Human coronavirus OC43-elicited CD4+ T cells protect against SARS-CoV-2 in HLA transgenic mice
Nature Communications (2024)
-
Distinctive evolution of alveolar T cell responses is associated with clinical outcomes in unvaccinated patients with SARS-CoV-2 pneumonia
Nature Immunology (2024)
-
Long-term humoral and cellular immunity after primary SARS-CoV-2 infection: a 20-month longitudinal study
BMC Immunology (2023)