Abstract
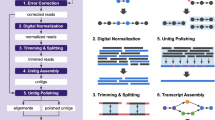
Massively parallel sequencing of cDNA has enabled deep and efficient probing of transcriptomes. Current approaches for transcript reconstruction from such data often rely on aligning reads to a reference genome, and are thus unsuitable for samples with a partial or missing reference genome. Here we present the Trinity method for de novo assembly of full-length transcripts and evaluate it on samples from fission yeast, mouse and whitefly, whose reference genome is not yet available. By efficiently constructing and analyzing sets of de Bruijn graphs, Trinity fully reconstructs a large fraction of transcripts, including alternatively spliced isoforms and transcripts from recently duplicated genes. Compared with other de novo transcriptome assemblers, Trinity recovers more full-length transcripts across a broad range of expression levels, with a sensitivity similar to methods that rely on genome alignments. Our approach provides a unified solution for transcriptome reconstruction in any sample, especially in the absence of a reference genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Birol, I. et al. De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Guttman, M. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510 (2010).
Haas, B.J. & Zody, M.C. Advancing RNA-Seq analysis. Nat. Biotechnol. 28, 421–423 (2010).
Yassour, M. et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl. Acad. Sci. USA 106, 3264–3269 (2009).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
De Bruijn, N.G. A combinatorical problem. Koninklijke Nederlandse Akademie v. Wetenschappen 46, 758–764 (1946).
Good, I.J. Normal recurring decimals. J. Lond. Math. Soc. 21, 167–169 (1946).
Pevzner, P.A., Tang, H. & Waterman, M.S. An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. USA 98, 9748–9753 (2001).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Butler, J. et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18, 810–820 (2008).
Hertz-Fowler, C. et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 32, D339–D343 (2004).
Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).
Parkhomchuk, D. et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123 (2009).
Rhind, N. et al. Comparative functional genomics of the fission yeasts. Science published online, doi:10.1126/science.1203357 (21 April 2011).
Wang, E.T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Wilhelm, B.T. et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243 (2008).
Xu, Z. et al. Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037 (2009).
Wu, T.D. & Watanabe, C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Wu, C.H. et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34, D187–D191 (2006).
Wapinski, I., Pfeffer, A., Friedman, N. & Regev, A. Natural history and evolutionary principles of gene duplication in fungi. Nature 449, 54–61 (2007).
Molnar, M. et al. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157, 519–532 (2001).
Nakamura, T., Kishida, M. & Shimoda, C. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells 5, 463–479 (2000).
Watanabe, T. et al. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29, 2327–2337 (2001).
Yassour, M. et al. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 11, R87 (2010).
Matlin, A.J., Clark, F. & Smith, C.W.J. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6, 386–398 (2005).
Robertson, G. et al. De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912 (2010).
Graveley, B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17, 100–107 (2001).
Wang, X.-W. et al. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics 11, 400 (2010).
Salzberg, S.L. & Yorke, J.A. Beware of mis-assembled genomes. Bioinformatics 21, 4320–4321 (2005).
Shannon, C.E. Prediction and entropy of printed English. Bell Syst. Tech. J. 30, 50–64 (1951).
Price, A.L., Jones, N.C. & Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 21 Suppl 1, i351–i358 (2005).
Grabherr, M.G. et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics 26, 1145–1151 (2010).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Kent, W.J. BLAT–the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Acknowledgements
We thank L. Gaffney for help with figure preparation, J. Bochicchio for project management, the Broad Sequencing Platform for all sequencing work, A. Papanicolaou and M. Ott for Inchworm software testing and code enhancements, and F. Ribeiro for helpful discussions regarding error pruning. The work was supported in part by a grant from the National Human Genome Research Institute (NIH 1 U54 HG03067, Lander), the Howard Hughes Medical Institute, a National Institutes of Health PIONEER award, a Burroughs Wellcome Fund–Career Award at the Scientific Interface (A.R.), the US-Israel Binational Science Foundation (N.F. and A.R.), and funds from the National Institute of Allergy and Infectious Diseases under contract no. HHSN27220090018C. M.Y. was supported by a Clore Fellowship. K.L.-T. is a recipient of the European Young Investigator Award (EYRYI) funded by the European Science Foundation. A.R. is a researcher of the Merkin Foundation for Stem Cell Research at the Broad Institute.
Author information
Authors and Affiliations
Contributions
M.G.G., M.Y., B.J.H., K.L.-T., N.F. and A.R. conceived and designed the study. B.J.H., M.G.G. and M.Y. developed the Inchworm, Chrysalis and Butterfly components, respectively. N.R., F.D.P., B.W.B., C.N., K.L.-T. contributed to the study's conception and execution. J.Z.L., D.A.T., X.A., L.F., R.R., I.A., N.H., A.R. and A.G. designed and performed all experiments. Q.Z., Z.C. and E.M. contributed computational analyses. M.G.G., B.J.H. and M.Y. designed, implemented and evaluated all methods. A.R., N.F., M.G.G., B.J.H. and M.Y. wrote the manuscript, with input from all authors. A.R. and N.F. contributed equally to this paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Methods, Supplementary Note and Supplementary Figures 1–9 (PDF 394 kb)
Rights and permissions
About this article
Cite this article
Grabherr, M., Haas, B., Yassour, M. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–652 (2011). https://doi.org/10.1038/nbt.1883
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1883
This article is cited by
-
Girdling behavior of the longhorn beetle modulates the host plant to enhance larval performance
BMC Ecology and Evolution (2024)
-
Functional similarity, despite taxonomical divergence in the millipede gut microbiota, points to a common trophic strategy
Microbiome (2024)
-
GTax: improving de novo transcriptome assembly by removing foreign RNA contamination
Genome Biology (2024)
-
Pangenome analysis reveals transposon-driven genome evolution in cotton
BMC Biology (2024)
-
Hooked on zombie worms? Genetic blueprints of bristle formation in Osedax japonicus (Annelida)
EvoDevo (2024)