Abstract
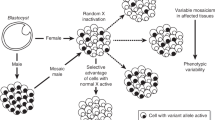
In female mammals, most genes on one X chromosome are silenced as a result of X-chromosome inactivation1,2. However, some genes escape X-inactivation and are expressed from both the active and inactive X chromosome. Such genes are potential contributors to sexually dimorphic traits, to phenotypic variability among females heterozygous for X-linked conditions, and to clinical abnormalities in patients with abnormal X chromosomes3. Here, we present a comprehensive X-inactivation profile of the human X chromosome, representing an estimated 95% of assayable genes in fibroblast-based test systems4,5. In total, about 15% of X-linked genes escape inactivation to some degree, and the proportion of genes escaping inactivation differs dramatically between different regions of the X chromosome, reflecting the evolutionary history of the sex chromosomes. An additional 10% of X-linked genes show variable patterns of inactivation and are expressed to different extents from some inactive X chromosomes. This suggests a remarkable and previously unsuspected degree of expression heterogeneity among females.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lyon, M. F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961)
Plath, K., Mlynarczyk-Evans, S., Nusinow, D. A. & Panning, B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 36, 233–278 (2002)
Willard, H. F. in The Metabolic and Molecular Bases of Inherited Disease 8th edn (eds Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B. & Vogelstein, B.) 1191–1221 (McGraw-Hill, New York, 2000)
Carrel, L., Cottle, A. A., Goglin, K. C. & Willard, H. F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl Acad. Sci. USA 96, 14440–14444 (1999)
Carrel, L. & Willard, H. F. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc. Natl Acad. Sci. USA 96, 7364–7369 (1999)
Brown, C. J. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44 (1991)
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S. & Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996)
Rougeulle, C. et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell. Biol. 24, 5475–5484 (2004)
Chadwick, B. P. & Willard, H. F. Barring gene expression after XIST: maintaining facultative heterochromatin on the inactive X. Semin. Cell Dev. Biol. 14, 359–367 (2003)
Chadwick, B. P. & Willard, H. F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl Acad. Sci. USA 101, 17450–17455 (2004)
Lyon, M. F. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 14, 135–148 (1962)
Brown, C. J. & Greally, J. M. A stain upon the silence: genes escaping X inactivation. Trends Genet. 19, 432–438 (2003)
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature doi:10.1038/nature03440 (this issue).
Miller, A. P. & Willard, H. F. Chromosomal basis of X chromosome inactivation: identification of a multigene domain in Xp11.21-p11.22 that escapes X inactivation. Proc. Natl Acad. Sci. USA 95, 8709–8714 (1998)
Huynh, K. D. & Lee, J. T. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426, 857–862 (2003)
Ohno, S. Sex Chromosomes and Sex-linked Genes (Springer, Berlin, 1967)
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003)
Graves, J. A. Mammals that break the rules: genetics of marsupials and monotremes. Annu. Rev. Genet. 30, 233–260 (1996)
Lahn, B. T. & Page, D. C. Four evolutionary strata on the human X chromosome. Science 286, 964–967 (1999)
Jegalian, K. & Page, D. C. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 394, 776–780 (1998)
Kohn, M., Kehrer-Sawatzki, H., Vogel, W., Graves, J. A. & Hameister, H. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 20, 598–603 (2004)
Gartler, S. M. & Riggs, A. D. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190 (1983)
Lyon, M. F. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80, 133–137 (1998)
Bailey, J. A., Carrel, L., Chakravarti, A. & Eichler, E. E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl Acad. Sci. USA 97, 6634–6639 (2000)
Mohandas, T., Sparkes, R. S. & Shapiro, L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science 211, 393–396 (1981)
Ke, X. & Collins, A. CpG islands in human X-inactivation. Ann. Hum. Genet. 67, 242–249 (2003)
Committee on Understanding the Biology of Sex and Gender Differences. In Exploring the Biological Contributions to Human Health–Does Sex Matter? (eds Wizemann, T. M. & Pardue, M.) (National Acadamies Press, Washington, DC, 2001)
Iyer, V. R. et al. The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87 (1999)
Esposito, T. et al. Escape from X inactivation of two new genes associated with DXS6974E and DXS7020E. Genomics 43, 183–190 (1997)
Acknowledgements
We thank M. Ross for sharing information before publication and gratefully acknowledge technical assistance from G. Nickel, K. Trevarthen, J. Dunn, A. Cottle and M. Moon. This work was supported in part by a National Institutes of Health research grant to H.F.W. and L.C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure S1
Y homology influences Xi expression. (DOC 23 kb)
Supplementary Figure S2
Xi expression data correlate with repetitive element composition. (DOC 23 kb)
Supplementary Figure S3
Gene organization and sequence composition of 500 kb regions on the X; repetitive elements cannot fully explain Xi expression patterns. (PDF 107 kb)
Supplementary Figure S4
Distribution of transcripts with CpG islands. (DOC 23 kb)
Supplementary Figure S5
Levels of relative Xa and Xi expression. (DOC 63 kb)
Supplementary Figure S6
RT-PCR assays of gene expression from the Xi chromosome. (DOC 357 kb)
Supplementary Table S1
Genes assayed by quantitative analysis of expressed polymorphisms in panel of non-randomly inactivated primary fibroblasts. (XLS 40 kb)
Supplementary Table S2
X-linked genes analyzed in this study. (XLS 16 kb)
Supplementary Table S3
Genes analysed in Xi hybrids. (XLS 476 kb)
Supplementary Table S4
Comparison of X inactivation patterns as measured in primary fibroblasts and in Xi hybrids. (XLS 16 kb)
Supplementary Note S1
Notes as to why some reported genes were not fully analysed. (DOC 23 kb)
Supplementary Note S2
Although the RT-PCR assay utilized is not strictly quantitative, substantial effort was made to ensure that Xi hybrids scored positive did have significant levels of expression. (DOC 20 kb)
Supplementary Note S3
The S3/S4 boundary has been assigned to a 2.5 Mb region within Xp22.2-Xp22.31. (DOC 19 kb)
Rights and permissions
About this article
Cite this article
Carrel, L., Willard, H. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005). https://doi.org/10.1038/nature03479
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03479
This article is cited by
-
Overcoming genetic and cellular complexity to study the pathophysiology of X-linked intellectual disabilities
Journal of Neurodevelopmental Disorders (2024)
-
Role of sex in immune response and epigenetic mechanisms
Epigenetics & Chromatin (2024)
-
YY1 binding is a gene-intrinsic barrier to Xist-mediated gene silencing
EMBO Reports (2024)
-
Genetic analysis of the X chromosome in people with Lewy body dementia nominates new risk loci
npj Parkinson's Disease (2024)
-
Oral administration of a recombinant modified RBD antigen of SARS-CoV-2 as a possible immunostimulant for the care of COVID-19
Microbial Cell Factories (2024)